Method for improving erythrocin yield through inactivation saccharopolyspora erythraea SACE_3446 gene
A technology of Saccharopolyspora erythromycetes and erythromycin, applied in the field of genetic engineering, can solve the problem of less research on the regulatory genes of Saccharopolyspora erythromycetes
Inactive Publication Date: 2012-12-12
ANHUI UNIVERSITY
View PDF2 Cites 11 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
In 2007, Oliynyk et al. [7] reported the genome sequence of S. bld (SACE_2077)[6] and SACE_7040[8] regulatory gene research reports
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment Construction
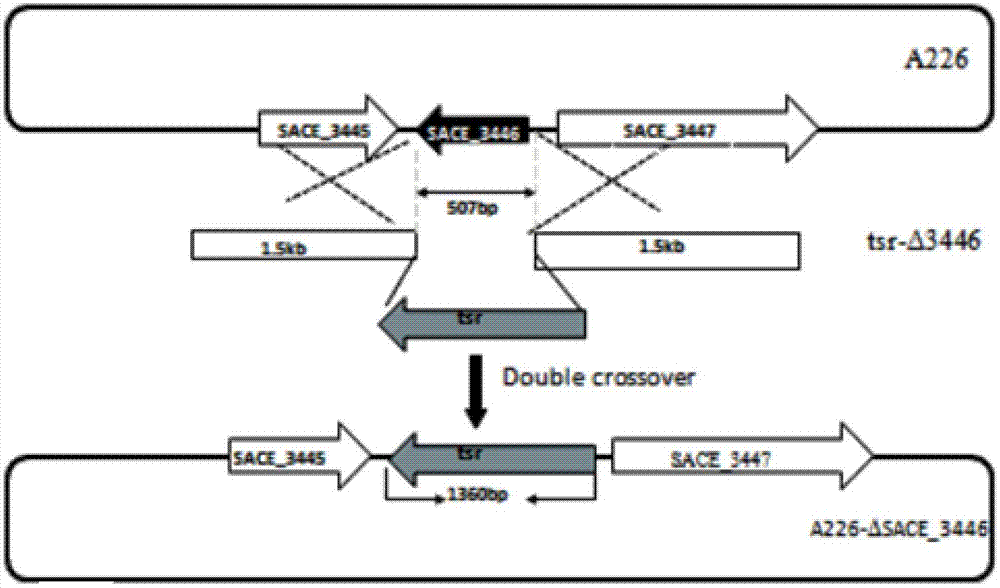
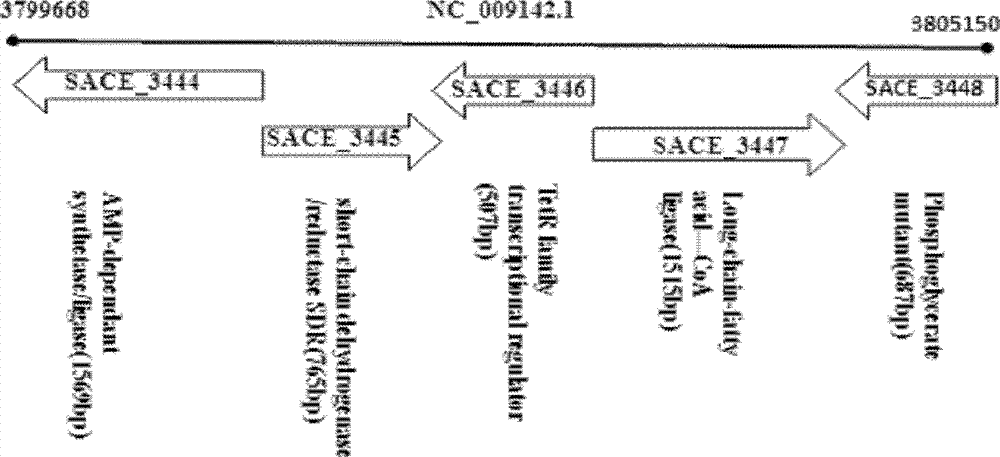
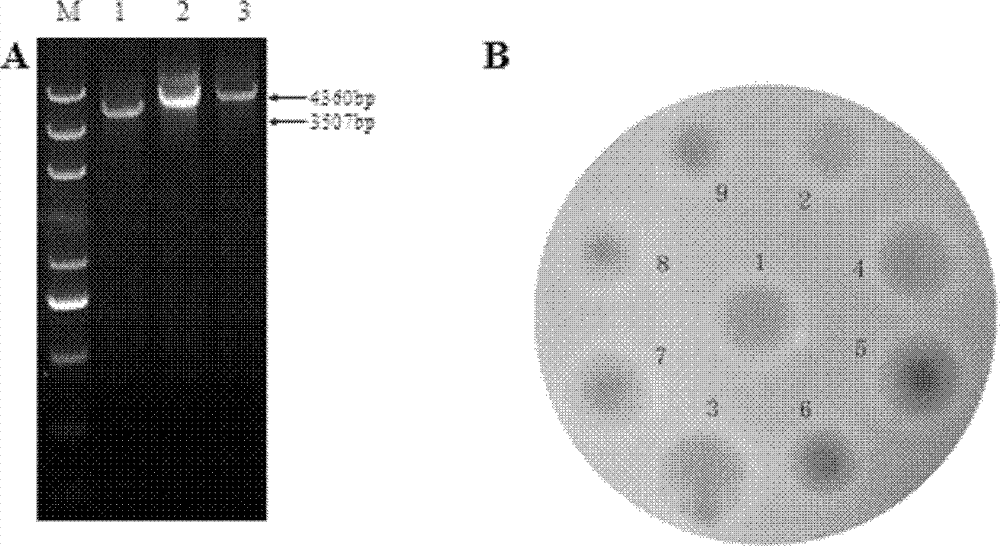
[0025] The embodiment of the present invention is not limited to the Saccharopolyspora Erythromyces A226 strain; the embodiment of the present invention uses but is not limited to the method of homologous recombination of chromosome fragments to obtain the gene inactivation or deletion of Saccharopolyspora Erythromyces SACE_3446.
[0026] Materials and Methods
[0027] 1.1 Strains, plasmids and growth conditions
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention discloses a method for improving erythrocin yield through a negative control gene SACE_3446 on an inactivation saccharopolyspora erythraea chromosome. Saccharopolyspora erythraea is used for producing erythrocin. The erythrocin and derived drugs of the erythrocin such as clarithromycin, azithromycin and telithromycin are used widely in clinic. Erythrocin high-producing strain screening is very important in industrial production. The erythromycin biosynthesis negative control gene SACE_3446 is screened from a saccharopolyspora erythraea TetR family. Compared with erythrocin yield of an original strain, deletion mutants of the saccharopolyspora erythraea SACE_3446 is improved remarkably, the erythrocin is returned to low yield after gene complementation of the SACE_3446, and therefore the SACE_3446 gene is a erythromycin biosynthesis negative control gene. The inactivation saccharopolyspora erythraea SACE_3446 gene can improve the erythrocin yield through a genetic engineering way. Due to the fact that erythromycin biosynthesis gene control is a network system, upstream and downstream control factors acted by SACE_3446 control factors can be found by using the SACE_3446 as an object. The erythrocin yield can also be improved by changing upstream or downstream control factor genes of the saccharopolyspora erythraea SACE_3446 control factors.
Description
technical field [0001] The invention belongs to the technical field of genetic engineering, and specifically constructs a genetic engineering strain by inactivating or deleting the erythromycin biosynthesis negative regulation gene SACE_3446 on the chromosome of Erythromycetes saccharopolyspora, and improves the yield of erythromycin by fermentation. Background technique [0002] Secondary metabolites of actinomycetes have a wide range of uses, such as antibiotics, anticancer agents, immunomodulators, anthelmintics, insect control agents [1,2]. Among the 23,000 biologically active secondary metabolites discovered so far, more than 10,000 are produced by actinomycetes. However, the original production of these secondary metabolites is very low, and screening is required to obtain high-yielding strains for industrial production. In the past, industrial strains were mainly obtained by random physical or chemical mutagenesis [3,4]. Traditional random mutagenesis techniques ar...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More IPC IPC(8): C12P19/62C12N15/09C12R1/645
Inventor 张部昌朱林黄训端
Owner ANHUI UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com