Micro RNAs (ribonucleic acids) fluorescence detection probe
A technology for detecting probes and fluorescent probes, which is applied in the fields of molecular biology and nucleic acid chemistry, can solve the problems of complex probe clusters, high cost and difficulty in operation, complex system detection cross-interference, etc., and achieve good discrimination and good The effect of response
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
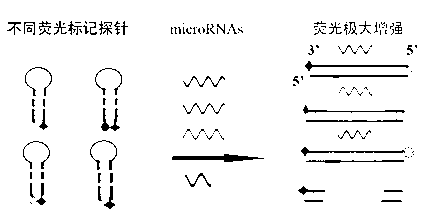
[0038] 1. Design of multicolor microRNAs detection probe system
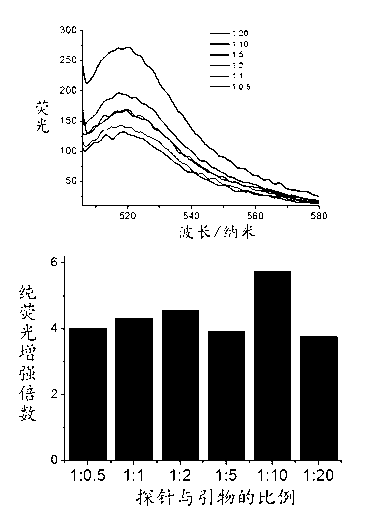
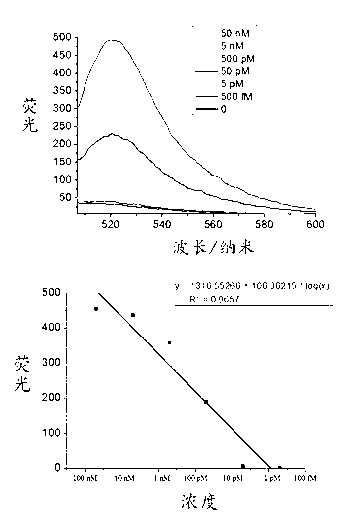
[0039] (1) The probe contains a region complementary to the target microRNA, which is responsible for recognizing the corresponding microRNA, and this part of the sequence is designed to specifically detect the target microRNA; the terminal region sequence, when there is no microRNA, the terminal region ensures that the probe has a folded structure, Such as figure 1 shown. The 5' end of the probe is labeled with different fluorophores, and the 3' end is marked with a quencher group. Under normal conditions, the probe is a folded structure, in which the distance between the fluorophore and the quencher group is close, and the fluorescence is quenched.
[0040] (2) The detection target to be achieved: It can produce a good signal response to the corresponding target microRNA probe, the fluorescence should be able to achieve obvious differences, and there should be no background for non-target microRNA.
[0041] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com