Anti-ustilaginoidea virens major gene and molecular marker thereof
A technology of molecular markers and rice false smut, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve problems such as gene linkage, achieve the effect of reducing breeding scale and improving breeding efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0019] The present invention will be further described below in conjunction with specific implementation examples. The following examples are only used to illustrate the present invention and are not intended to limit the scope of protection of the present invention. Without departing from the essence of the present invention, any replacement of the methods, steps or recipient parents of the present invention falls within the scope of the present invention. The experimental methods in the following implementation examples are conventional methods unless otherwise specified.
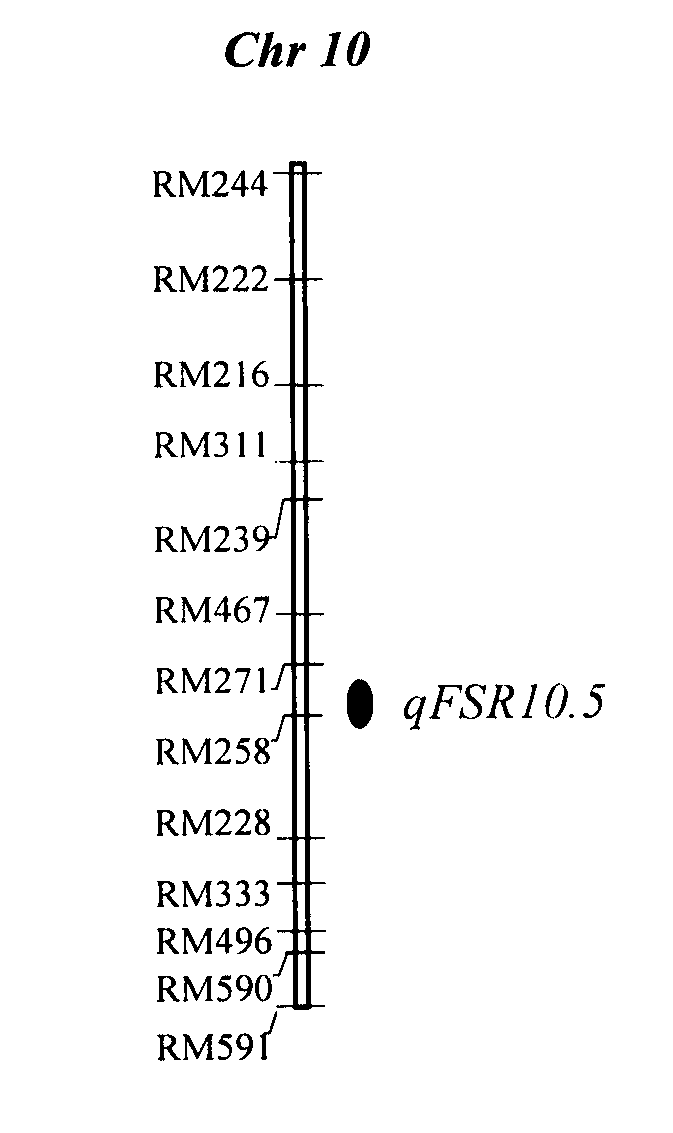
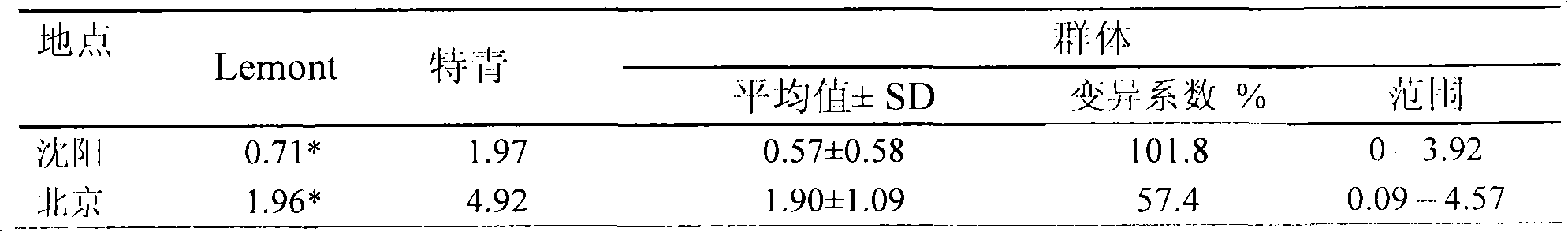
[0020] 1. Positioning population and identification of resistance
[0021] Teqing background backcross introduction line population consists of 213 lines, including 133 BC 2 f 5 and 80 BC 3 f 4 strain. Teqing is a high-yield indica rice variety widely promoted in my country, and Lemont is a high-quality, high-yield japonica rice variety popularized in the southern United States.
[0022] In the ric...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com