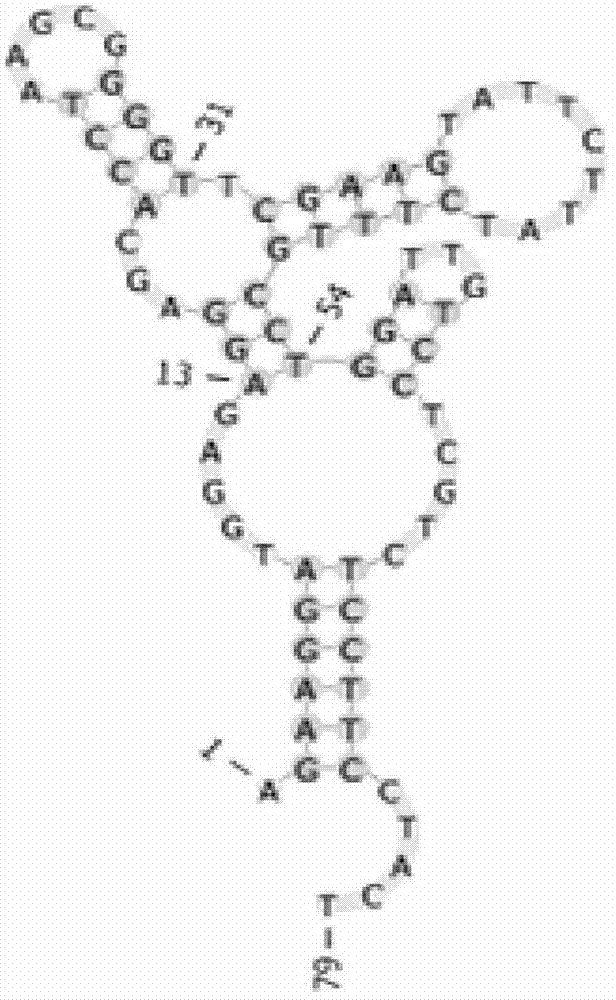Nucleic acid aptamer and derivatives thereof, screening method of nucleic acid aptamer, application of nucleic acid aptamer and derivatives in detecting human biliary duct carcinoma cell line
A technology of nucleic acid aptamer and screening method, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, DNA preparation, etc. good effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
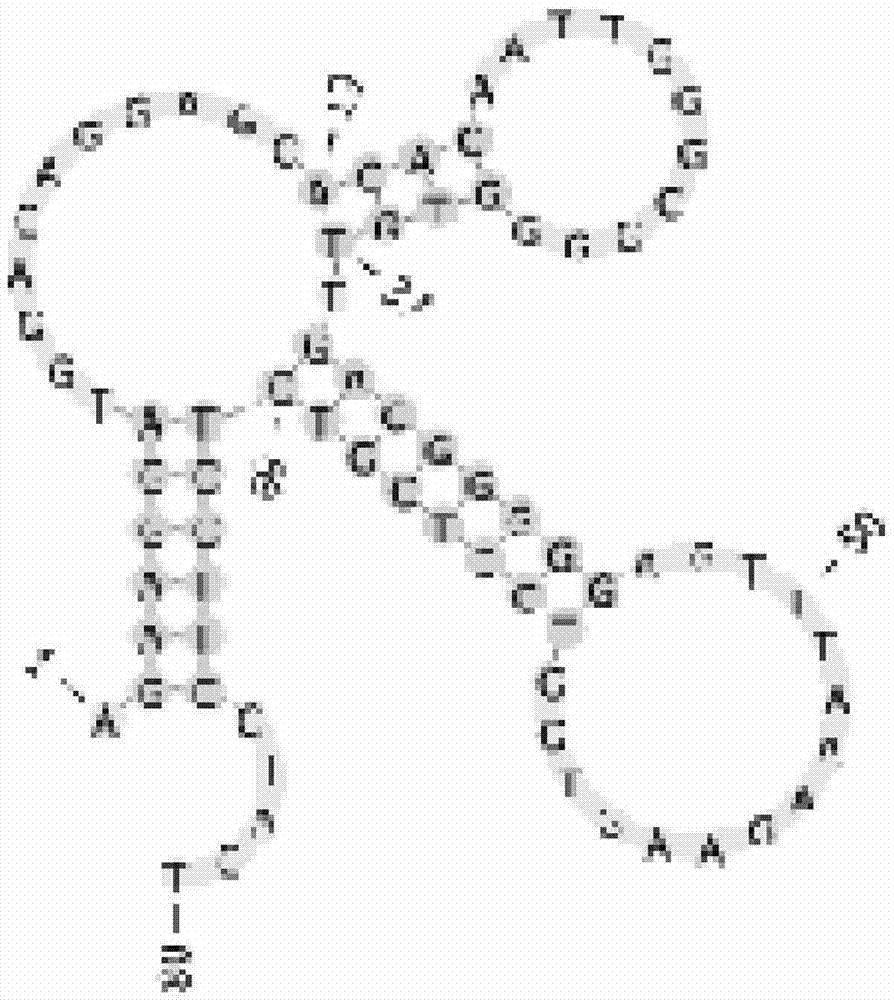
[0046] A nucleic acid aptamer that can be used to detect the human hilar cholangiocarcinoma cell line QBC-939, the nucleotide sequence of the nucleic acid aptamer includes the DNA fragment shown in any one of the following sequences 1 to 2 ( The 5' end is labeled with FAM):
[0047] Sequence 1FAM-yl19:
[0048] 5'-AGAAGGATGGAGAGGAGCACACAATTGGGCGGGGTGTTGACGGGGGAGTTTAAAG
[0049] AAGTGGTCCTCGTCTCCTTCCTACT-3';
[0050] Sequence 2FAM-yl23:
[0051] 5'-AGAAGGATGGAGAGGAGCACCTAAGCGGGGTTCGAAGTATTTCTTATCTTTGCCTGG
[0052] ATTGTCCTCGTCTCCTTCCTACT-3'.
[0053] Serial 3FAM-RS80:
[0054] 5'-NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
[0055] NNNNNNNNNNNNNNNNNNNNNNNNNNNNNN-3’
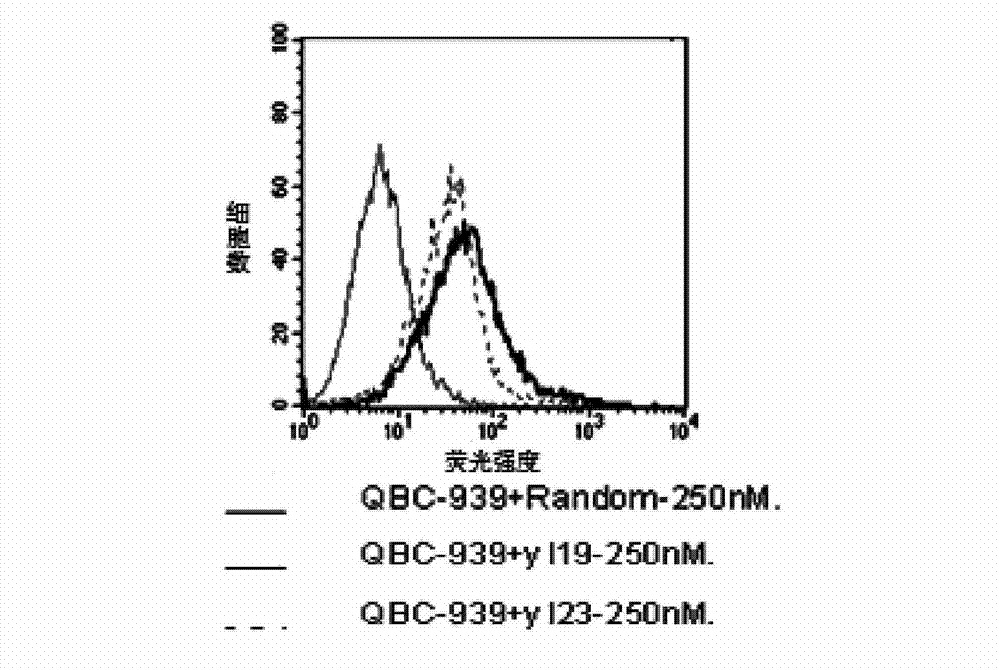
[0056] Note: N represents any base in A, T, G, and C (sequence 3 is used as a control sequence, that is, a negative probe, that is image 3 , Figure 4 Random in ).
[0057] A certain position on the nucleotide sequence of each nucleic acid aptamer mentioned above can be phosphorylated, methylate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com