Method for identifying bacteria microflora composition in fermented sourdough
A technology for fermenting sourdough and bacteria, which is applied in biochemical equipment and methods, determination/inspection of microorganisms, etc., can solve problems such as being in the primary stage, and achieve the effect of low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
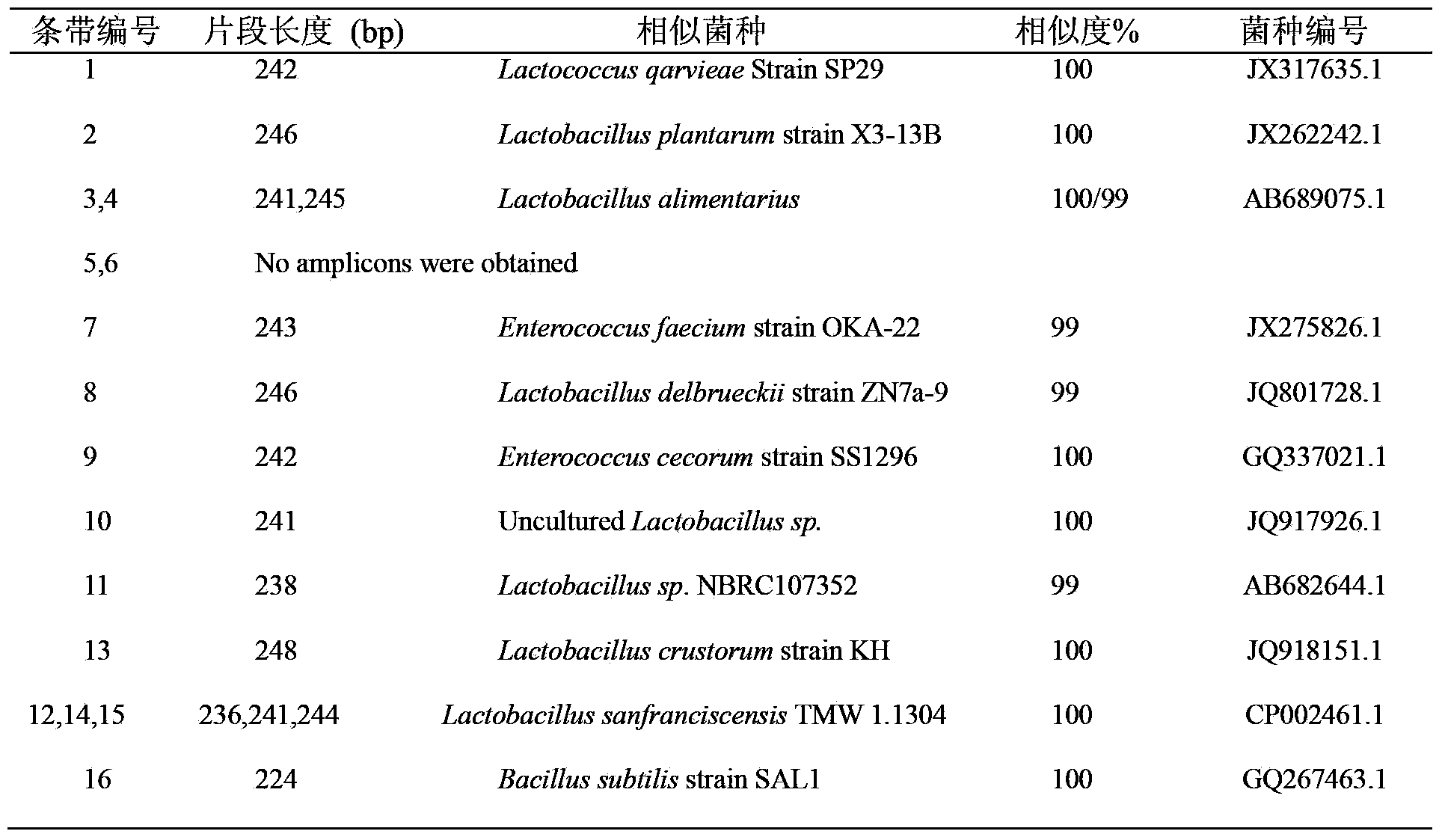
Image
Examples
Embodiment 1
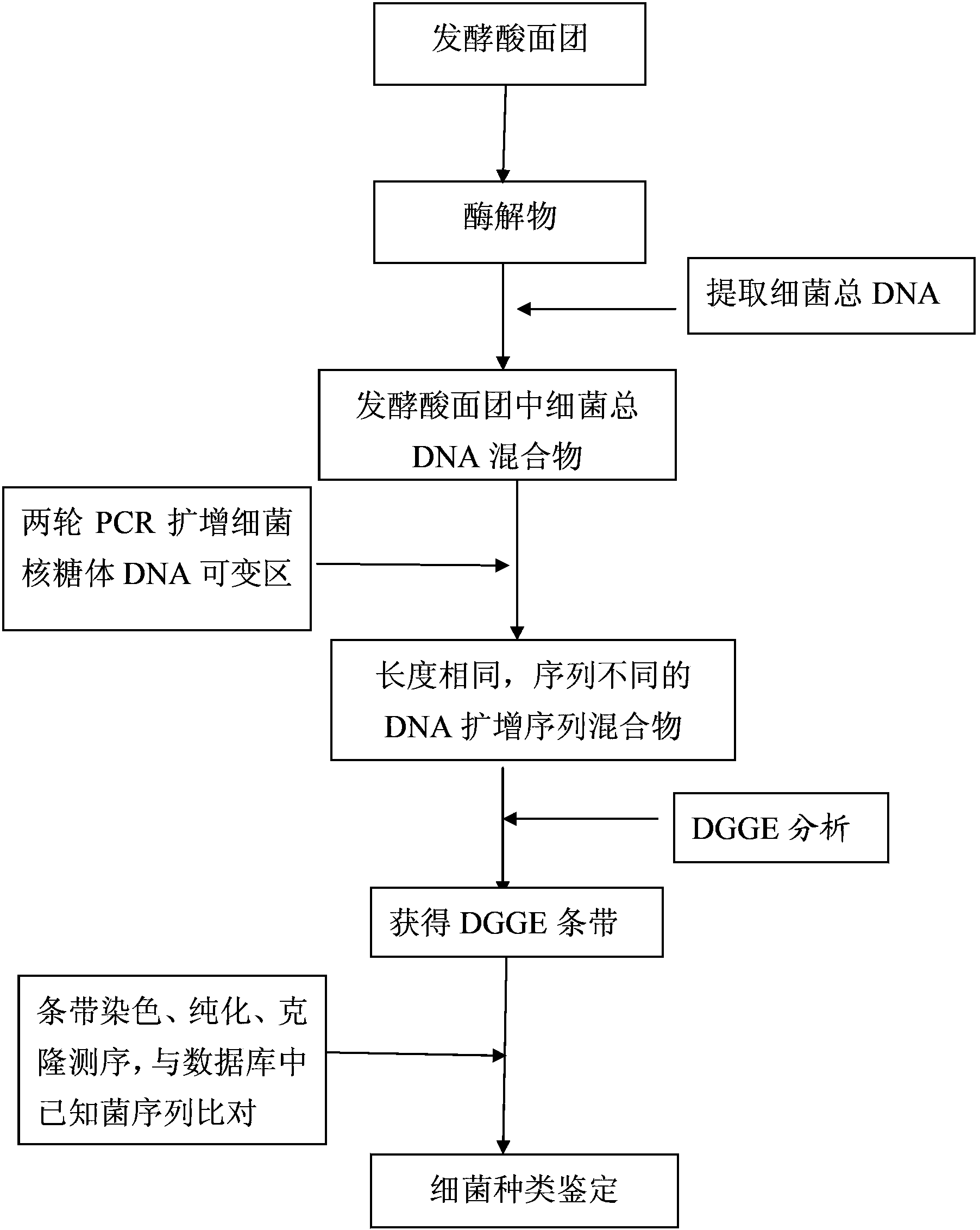
[0039] as per figure 1 The technical process shown in the operation is as follows:
[0040] (1) Take five traditional fermented sourdoughs from different regions in my country, numbered Hn-87, Sx-91, Gs-107, Hf-112 and Hr-122, and take 200mg of fermented sourdoughs respectively in 1.5mL centrifuge tubes , add 1mL of 20% α-amylase solution, shake and mix.
[0041] (2) Heat the mixture in a metal bath at 65°C for 20 minutes, centrifuge at 12,000×g for 5 minutes, discard the precipitate, and take the supernatant to obtain the hydrolyzate.
[0042] (3) Bacterial DNA OMEGA kit was used to extract the total bacterial genomic DNA in the enzymatic hydrolyzate.
[0043] (4) Nandrop2000 detects the concentration of the extracted DNA, and adjusts the DNA concentration to 50ng / uL.
[0044] (5) Perform touchdown PCR (Touchdown PCR, the first round of PCR) amplification of the adjusted DNA solution;
[0045] The primers are bacterial universal primers 16S rDNA V3 region 341f and 543r wit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com