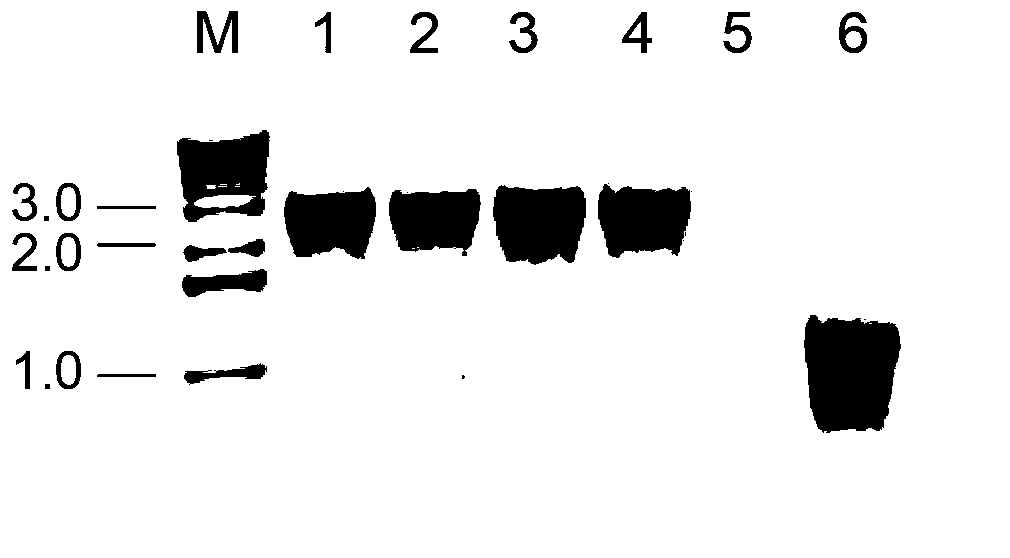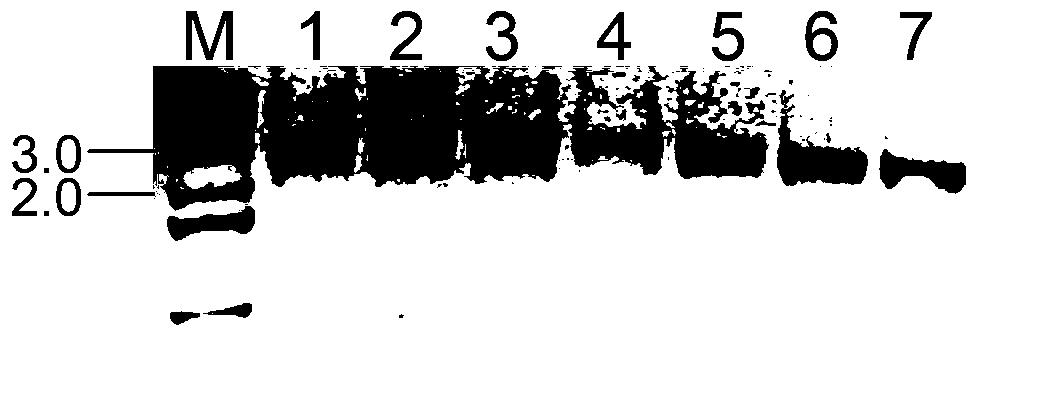Method for knocking out ibuprofen fosmid catechol dioxygenase gene
A technology of catechol dioxygenase and forsmid, which is applied in the field of ibuprofen microbial degradation, can solve the problems of inability to promote DNA strand insertion and self-exchange, excessive electrical energy and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1: Confirmation of 3G7Tn5 mutants G9 and H5 cloned from the Fossmid library
[0081] (1) After using the MilliQ system to prepare pure water for the PCR reaction test, filter it again using a 0.2 μm sterile filter membrane for later use.
[0082] (2) The above sterile ddH 2 O was dispensed into eppendorf centrifuge tubes, sterilized under UV light for 10 min, and used as water for PCR reactions.
[0083] (3) Start PCR Master Mix buffers 10 μL, primer ipf23O F (5'-ATA ATC GCA CGG AAC TCG ACA CCT-3') and R (5'-TCG GTG CGA CCT TCA ATC ATG TCA-3') each 1 μL, make up dd H 2 0 to 20 μL. Add a small amount of single colony (3G7 Tn5 mutant G9 and 3G7 Tn5 mutant H5 respectively) to the PCR reaction tube, the PCR reaction tube without colonies was used as a negative control, and the PCR reaction tube added with EPI3003G7 single colony was used as a positive control.
[0084] (4) After the PCR reaction is over, add 5 μL of 1kb Marker and 10 μL of PCR reacti...
Embodiment 2
[0086] Electric shock transformation of embodiment two recombinant plasmid pKD46
[0087] First prepare the competent cells of the 4F6 strain, the specific operation is as follows:
[0088] (1) Transfer 600 μL of the overnight culture solution of the 4F6 strain to 5.4 mL of LB culture solution supplemented with chloramphenicol (see Table 1 for the amount of antibiotics added), and incubate at 37°C for 3 hours.
[0089] (2) Aliquot the above culture solution into 1.5mL centrifuge tubes, centrifuge at 14,000rpm for 1.5 minutes, discard the supernatant, and repeat the centrifugation twice.
[0090] (3) Wash the above cells 3 times with 1 mL of 300 mM frozen sucrose, and resuspend in 100 μL of frozen sucrose.
[0091] (4) Add 80 μL of competent cells and 1 μL of pKD46 plasmid to a frozen electroshock transformation cup (use an electroshock transformation cup with a width of 1 mm), select 1.8 kV electric shock, quickly add 600 μL of LB culture medium, and recover for 1...
Embodiment 3 G7
[0097] Electroporation Transformation of Linear DNA in Example 3 3G7Tn5 Mutant
[0098] (1) Use ipf23O F and R primers to amplify the linear DNA in the Tn5 mutants G9 and H5 of 3G7 respectively, and the PCR amplification steps are the same as in Example 1.
[0099] (2) Cut glue, Zymoclean TM Gel DNA Recovery Kit purified and recovered samples, and detected the linear DNA above on 1.0% (w / v) agarose gel.
[0100] Agarose gel detection of linear DNA in PCR-amplified 3G7Tn5 mutants G9 and H5 see figure 2 . The bands amplified from the three G9 templates and the four H5 templates were all between 2.0-3.0kb. and figure 1 Same result.
[0101] During the preparation of competent cells of the 4F6-pKD46 strain, 1000× copy number induction solution needs to be added and cultured for 1.5 hours, and the rest of the electroporation transformation method is the same as in Example 2, with 2 repetitions.
[0102] (3) The cells were spread on LB double-antibody plates su...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com