Site-directed modification method of rice genome
A genetic modification and rice technology, applied in the field of plant genetic engineering, can solve problems such as high cost, difficult technology, complicated operation process, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0051] 1. Preparation of Cas9 nuclease recombinant expression vector
[0052] The codon optimization of Streptococcus pyogenes Cas9 (SpCas9, Streptococcus pyogenes Cas9) gene was carried out, and the nuclear localization signal (NLS) and BamHI / MfeI restriction enzyme cutting sites were added at both ends of the gene coding sequence to make the optimized Cas9 Better expression and localization in rice. The nucleotide coding sequence of Cas9 after adding NLS and restriction sites at both ends and codon optimization is shown in SEQ ID No. 1 in the sequence listing. In SEQ ID No.: 1, the 1st-6th position is the BamHI restriction site, the 4213-4218th position is the MfeI restriction site, the 10th-36th position is the N-terminal NLS sequence, and the 4162-4209th position is the C-terminal NLS sequence, the 37th-4161st position is the coding gene of Cas9 protein. The coding frame in SEQ ID No.: 1 is the 7th-4212th nucleotide of SEQ ID No.: 1, which encodes the protein shown in SE...
Embodiment 1
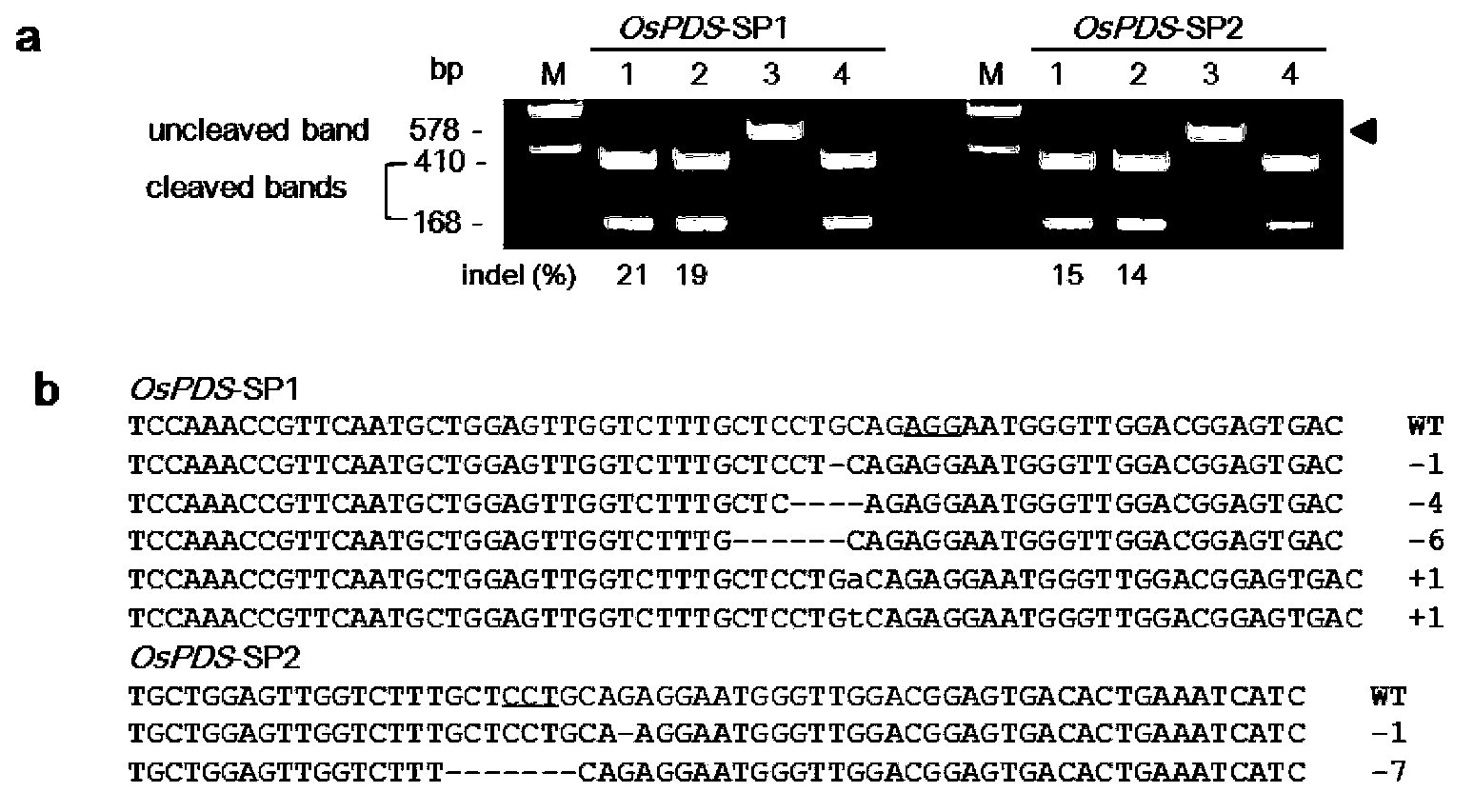
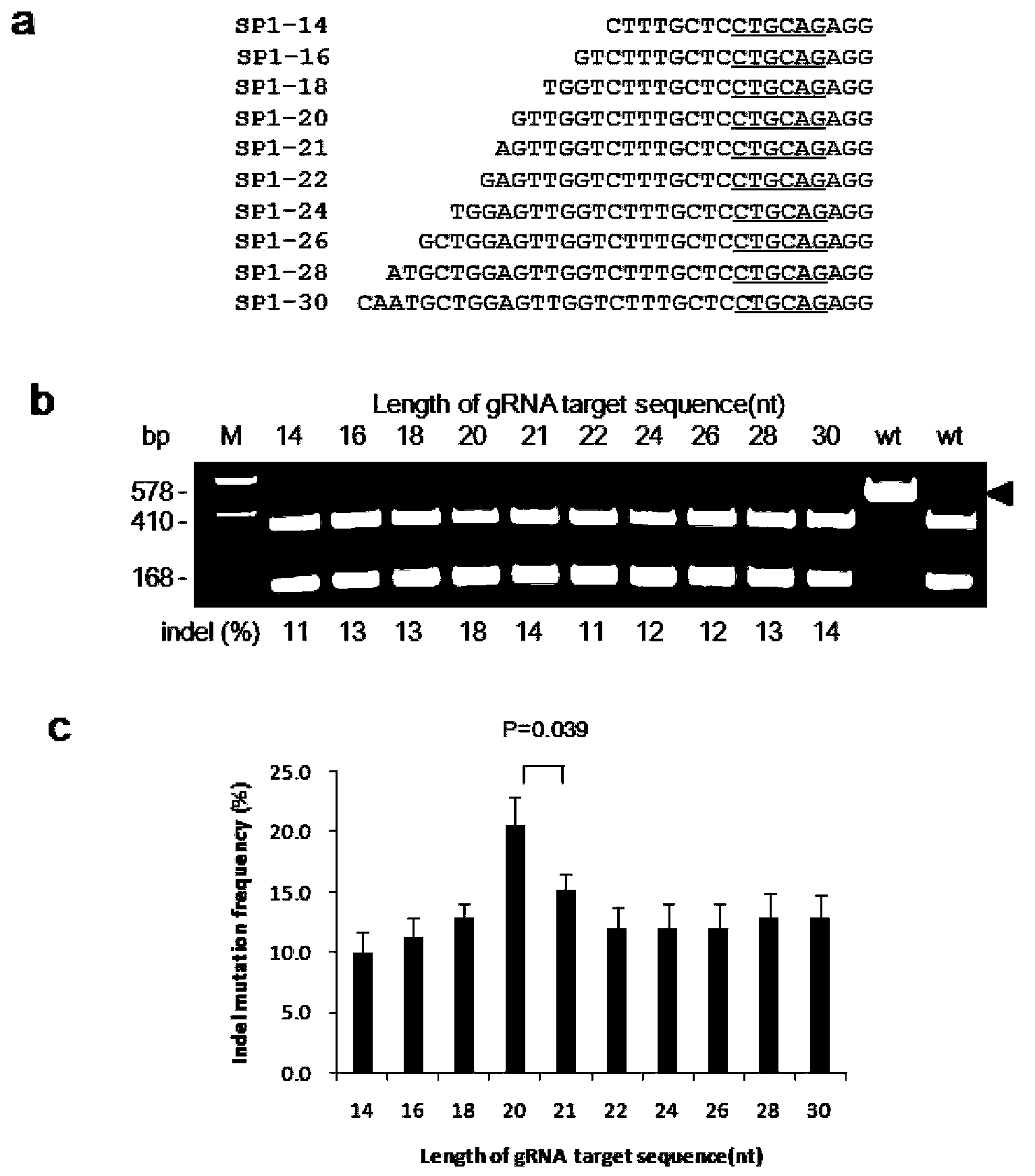
[0058] Example 1. Site-directed mutagenesis of rice endogenous gene OsPDS by gRNA:Cas9 system
[0059] (1) Design of target fragments target-SP1 and target-SP2
[0060] target-SP1: GTTGGTCTTTGCTCCTGCAG AGG ; (Locus No. 1353-1375 nucleotides in the OsPDS gene of LOC_Os03g08570.1.)
[0061] target-SP2:GTCCAACCCATTCCTCTGC AGG (Locus No. is the 1366th-1387th nucleotide of the negative strand in the OsPDS gene of LOC_Os03g08570.1.)
[0062] (2) Preparation of pU3-gRNA plasmids containing SP1 or SP2 nucleotide fragments
[0063] SP1 is the coding DNA of the RNA that can complementarily bind to the target target-SP1, and SP2 is the coding DNA of the RNA that can complementarily bind to the target target-SP2.
[0064] Synthesize the following single-stranded primers with sticky ends (underlined):
[0065] SP1-F: GGCA GTTGGTCTTTTGCTCCTGCAG,
[0066] SP1-R: AAAC CTGCAGGAGCAAAGACCAAC;
[0067] SP2-F: GGCA GTCCAACCCATTCCTCTGC,
[0068] SP2-R: AAAC GCAGAGGAATGGGTTGGAC.
...
Embodiment 2
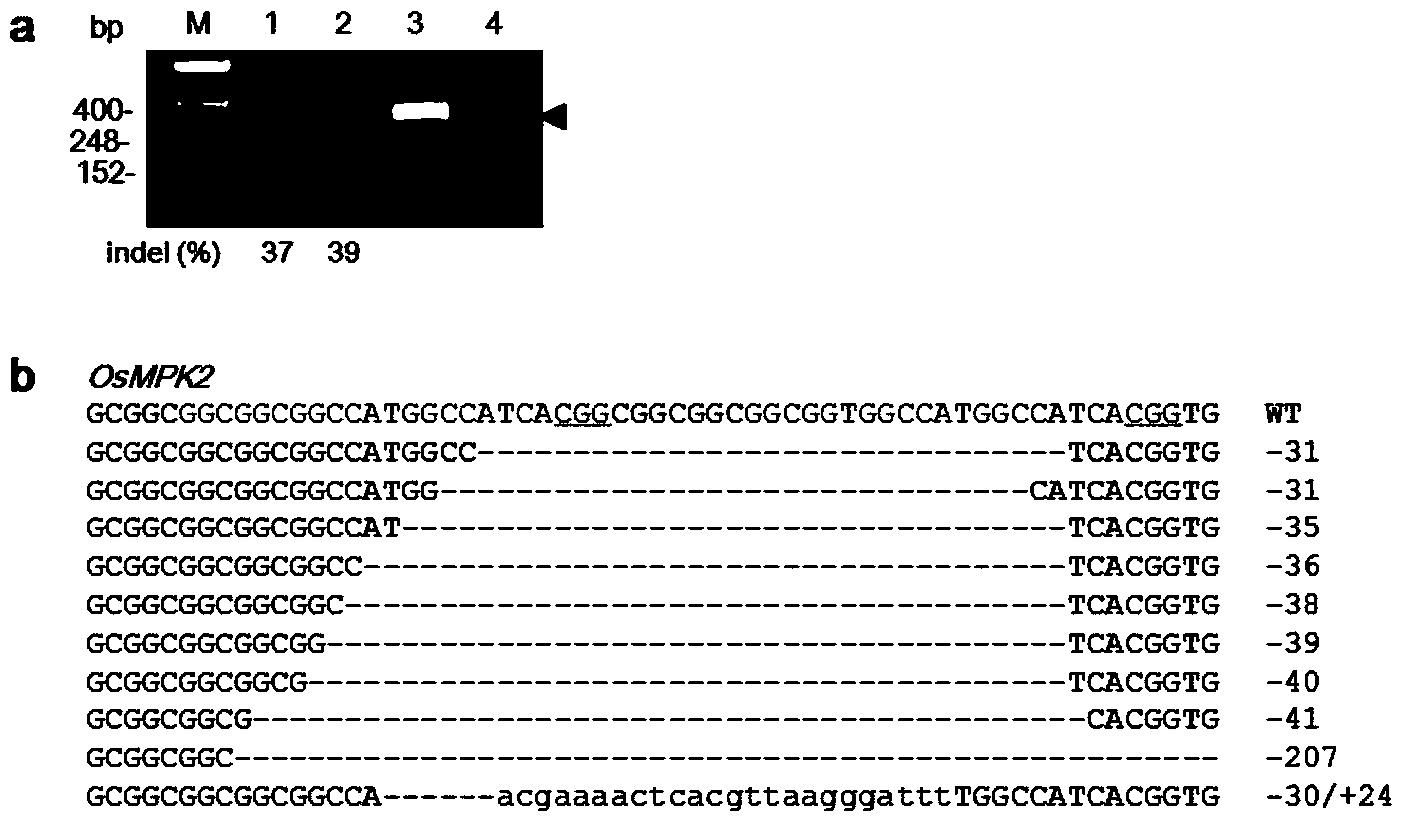
[0079] Example 2. Site-directed mutagenesis of rice endogenous genes OsBADH2 and OsMPK2 by gRNA: Cas9 system
[0080] (1) Design of target fragments target-SP3 and target-SP4
[0081] target-SP3: GCAGATCTTGCAGAATCCT TGG (Locus No. is 1785-1806 nucleotides in the OsBADH2 gene of LOC_Os08g32870.)
[0082] target-SP4: GCGGCGGCCATGGCCATCA CGG Please add (Locus No is 142-163 nucleotides in the OsMPK2 gene of LOC_Os08g06060.)
[0083] (2) Preparation of pU3-gRNA plasmids containing SP3 or SP4 nucleotide fragments
[0084] SP3 is the coding DNA of RNA that can complementarily bind to the target target-SP3, and SP4 is the coding DNA of RNA that can complementarily bind to the target target-SP4.
[0085] Synthesize the following single-stranded primers with sticky ends (underlined):
[0086] SP3-F: GGCA GCAGATCTTGCAGAATCCT,
[0087] SP3-R: AAAC AGGATTCTGCAAGATCTGC;
[0088] SP4-F: GGCAGCGGCGGCCATGGCCATCA,
[0089] SP4-R: AAAC TGATGGCCATGGCCGCCGC.
[0090] After the pri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com