Specific primer pairs used for auxiliary identification of potato viruses and application thereof
A specific primer pair, potato virus technology, applied in microorganisms, microorganism-based methods, biochemical equipment and methods, etc., can solve the problems of limitation, reduced sensitivity, time-consuming and laborious, and achieve good specificity and sensitivity. Widely popularized and applied , the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] The design of embodiment 1, potato virus primer
[0047] According to the conserved sequences of three potato viruses, 20 pairs of specific primers for potato virus X (PVX), 20 pairs of specific primers for potato virus S (PVS), and 18 pairs of specific primers for potato leafroll virus (PLRV) were designed. After screening and optimization, one pair of specific primers for PVX, PVS, and PLRV were obtained, as follows:
[0048] Specific primer pair for PVX (also known as specific primer pair A, the target sequence is 391bp):
[0049] Upstream primer F1: 5'-ACTGCAGGCGCAACTCCTGCCACAG-3' (sequence 1 of the sequence listing);
[0050] Downstream primer R1: 5'-GTGCTTGCCAGTTAGCAGGTGGAC-3' (SEQ ID NO: 2 in the Sequence Listing).
[0051] Specific primer pair for PVX (also known as specific primer pair B, the target sequence is 231bp):
[0052] Upstream primer F2: 5'-ATCGATGAGCTGTTCAAGATGG-3' (sequence 3 of the sequence listing);
[0053] Downstream primer R2: 5'-GATCGAGTCC...
Embodiment 2
[0057] Embodiment 2, the specificity of primer pair (singleplex RT-PCR)
[0058] 1. Specificity of specific primers to formazan
[0059] Potato virus X, potato S virus and potato leafroll virus are carried out as follows respectively:
[0060] 1. Extract the total RNA of the virus and reverse transcribe it into cDNA.
[0061] 2. Use the cDNA in step 1 as a template, and use specific primers to perform PCR amplification on A to obtain a PCR amplification product.
[0062] PCR amplification system: template 2 μl, 2×PCR buffer 10 μl, upstream primer (20 μM) 1 μl, downstream primer (20 μM) 1 μl, ddH 2 O 6 μl.
[0063] PCR amplification conditions: 94°C for 3min; 30 cycles of 94°C for 30s, 58°C for 30s, and 72°C for 30s; 72°C for 10min.
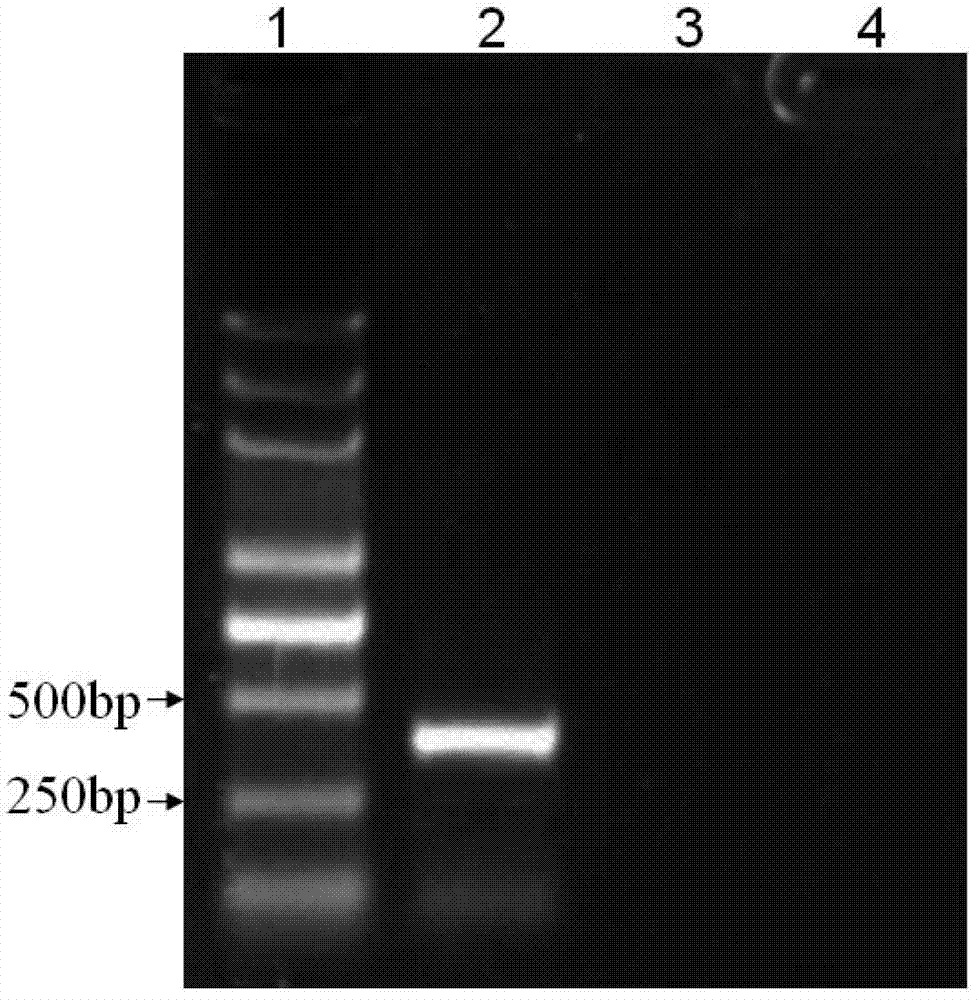
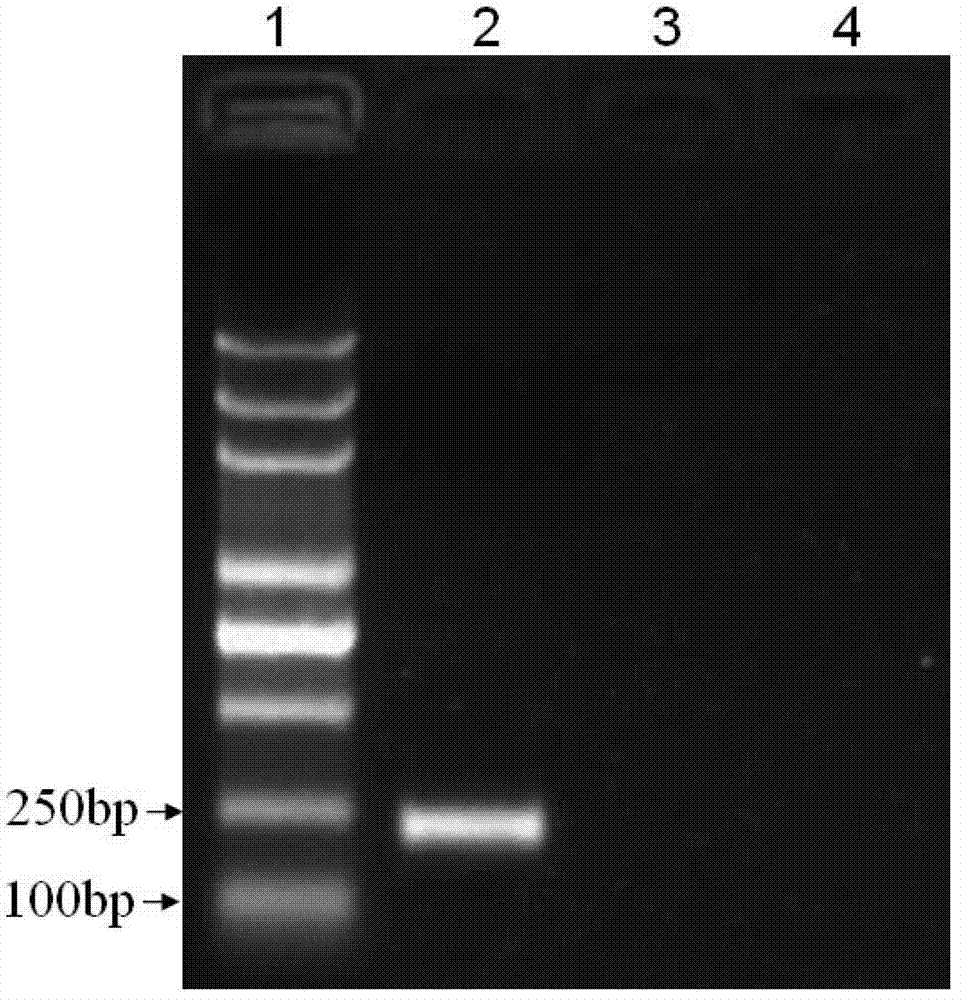
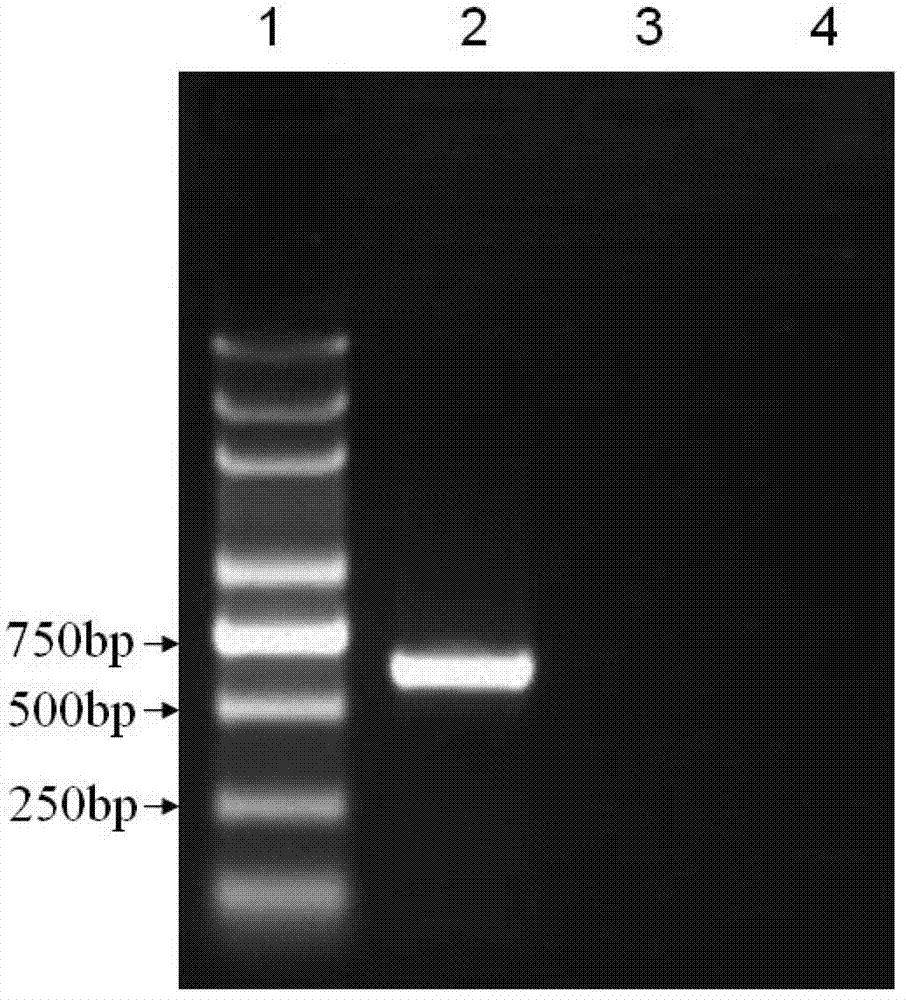
[0064] 3. Perform 1.5% agarose gel electrophoresis on the PCR amplification product, the results are shown in figure 1 ( figure 1 Among them, lane 1 is molecular weight marker, lane 2 is potato virus X, lane 3 is potato virus S, and lane 4 i...
Embodiment 3
[0077] Embodiment 3, the sensitivity of primer pair
[0078] 1. Sensitivity of specific primers to formazan
[0079] 1. Extract the total RNA of Potato virus X and reverse transcribe it into cDNA, and measure the cDNA concentration with a nucleic acid protein analyzer. After determination, the cDNA concentration of PVX is 501.5 ng / μl.
[0080] 2. Dilute the cDNA with sterile water to 400ng / μl, 300ng / μl, 200ng / μl, 100ng / μl, 50ng / μl, 25ng / μl and 10ng / μl with seven concentration gradient dilutions (according to the concentration from high to Low, named dilution 1 to dilution 7 in sequence).
[0081] 3. Using each dilution as a template, PCR amplification is carried out on armor with specific primers to obtain PCR amplification products.
[0082] The PCR amplification system and PCR amplification conditions are the same as Step 1 of Example 2.
[0083] 4. Perform 1.5% agarose gel electrophoresis on the PCR amplification product, see the results Figure 4 ( Figure 4 In, lane ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com