Prediction method and prediction device for protein side chain
A prediction method and protein technology, applied in the field of protein-related data processing, can solve the problems that deterministic algorithms cannot obtain optimal solutions, lack of universality, etc., and achieve the effects of stable accuracy, improved accuracy, and improved quality.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
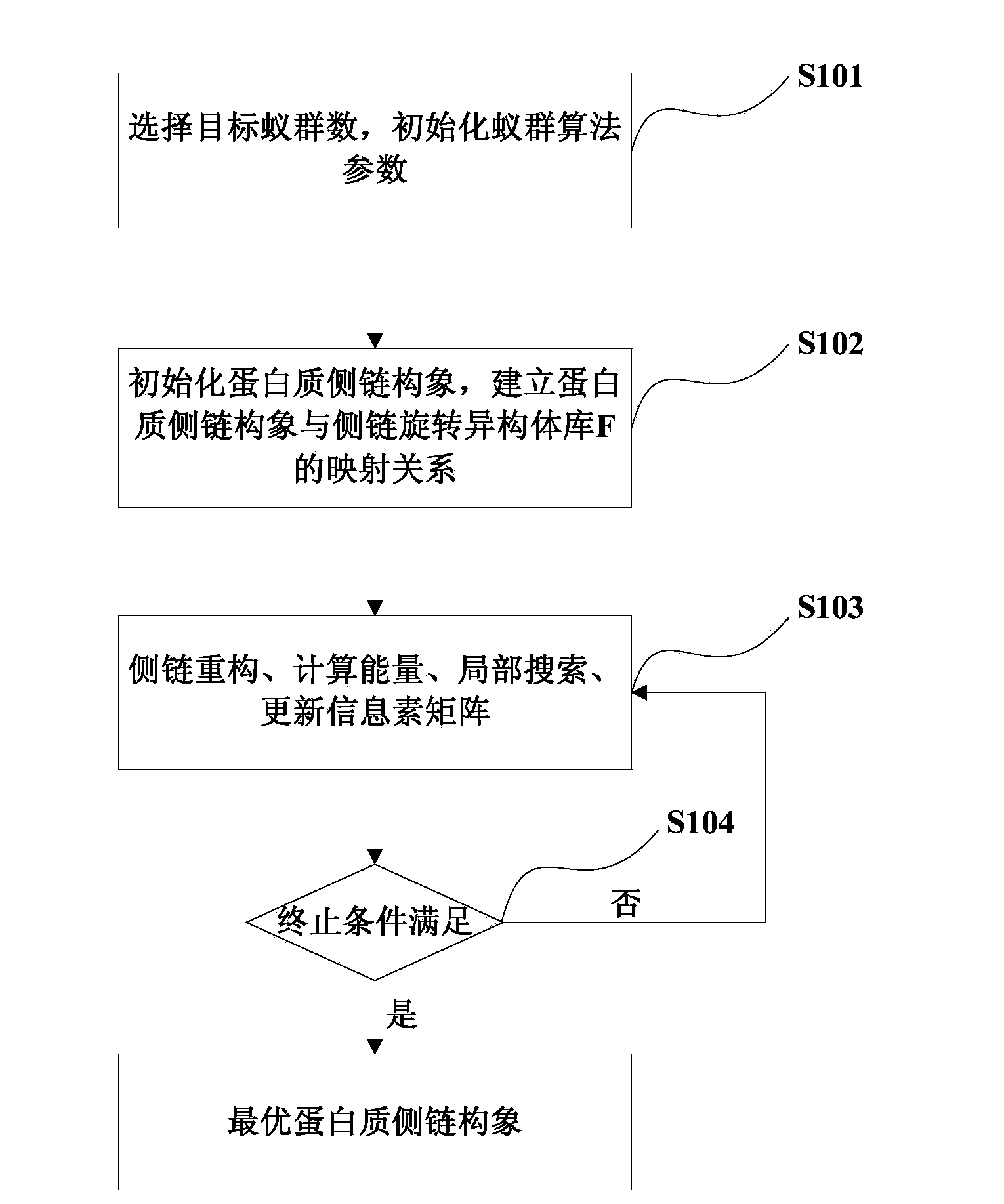
[0045] Embodiment one: see figure 1 As shown, it is a flow chart of the protein side chain prediction method provided in Embodiment 1 of the present invention.
[0046] Step S101, select the target ant colony, and initialize the parameters of the ant colony algorithm.
[0047] Step S101 is executed to select the target ant colony, that is, to determine the number of the target ant colony. In this embodiment, the number of ant colonies is determined to be 1, that is, to perform prediction based on the single ant colony algorithm.
[0048]Initialize the ant colony algorithm parameters, the ant colony algorithm parameters include:
[0049] The number m of ants is set to 30 in the embodiment of the present invention;
[0050] Used to determine the maximum number of cycles, which is set to 100 in this embodiment;
[0051] It is used to determine the algebra to be kept for the lowest energy side chain as the global optimal solution, which is set to 15 in this embodiment;
[0052...
Embodiment 2
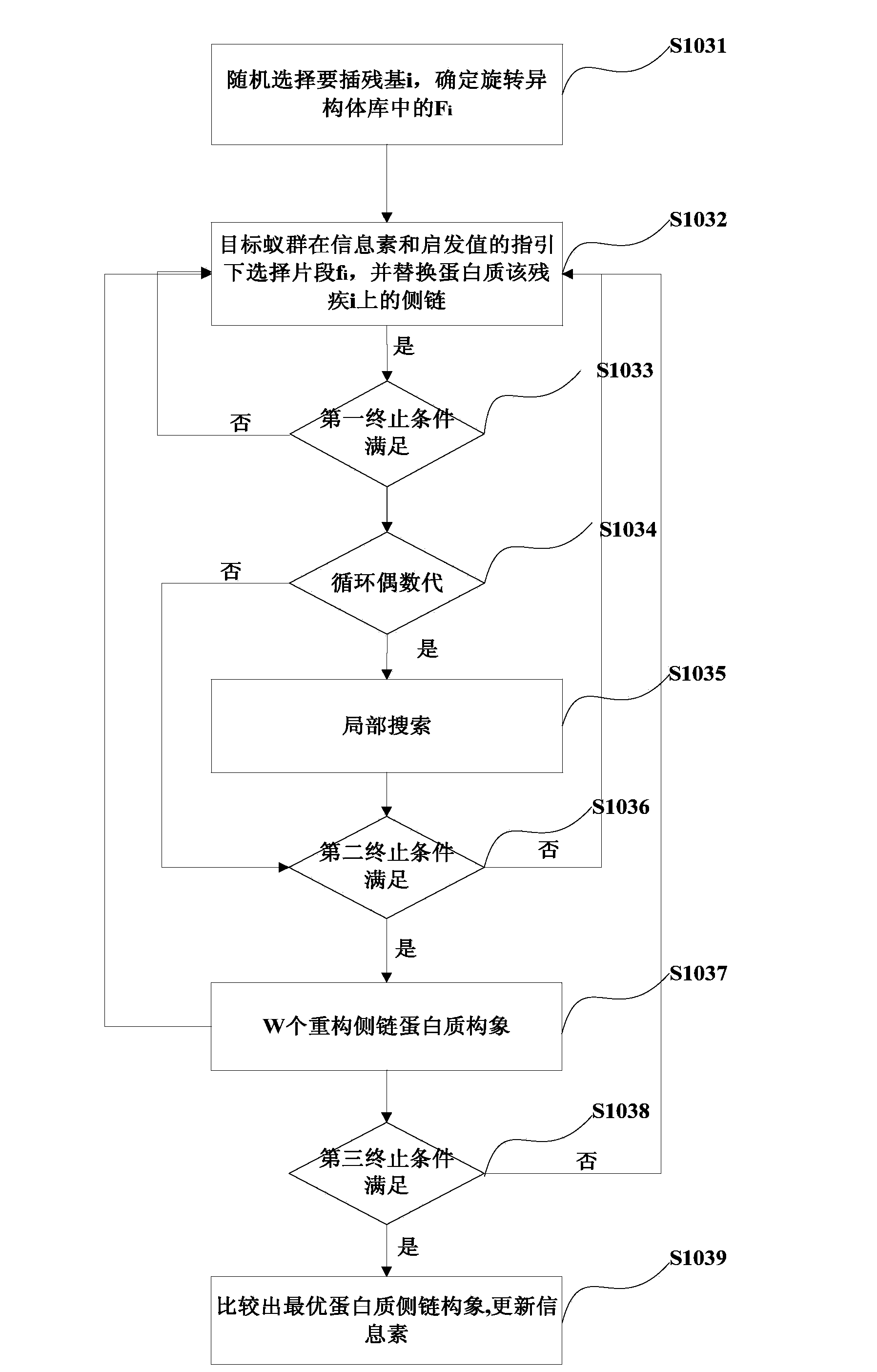
[0085] On the basis of the disclosure of the first embodiment, the present invention also discloses an embodiment, see Figure 4 , Figure 4 It is a flowchart of the method for predicting protein side chains based on parallel ant colony provided by Embodiment 2 of the present invention.
[0086] Different from Example 1, when selecting target ant colonies in Example 2, p ant colonies are selected, and p > 1, that is, in Example 2, multiple ant colonies are used to predict the protein structure in parallel. When selecting the target ant colony, the number of target ant colonies can be determined according to the number of processors, and in this embodiment, the number of ant colonies is determined to be 8.
[0087] When initializing the parameters of each ant colony algorithm, the 8 target ant colonies share the same pheromone matrix, that is, the 8 target ant colonies share the same pheromone matrix to predict the three-dimensional structure of the same protein.
[0088] In...
Embodiment 3
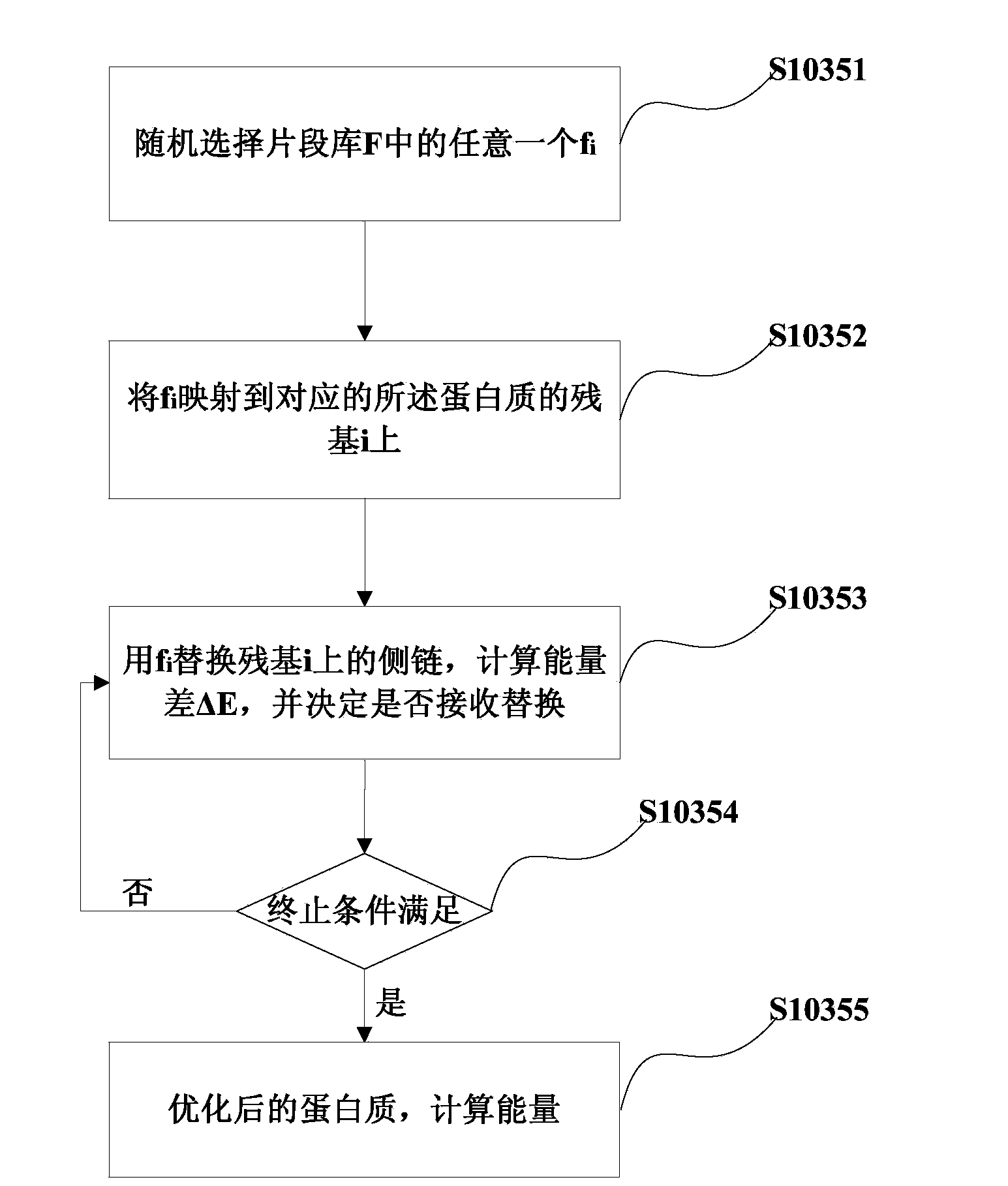
[0096] Embodiment 3 of the present invention discloses a device for predicting protein side chains based on ant colony algorithm, see Figure 5 , Figure 5 Schematic diagram of the structure of the protein side chain prediction device provided by the embodiment of the present invention.
[0097] The forecasting device mainly includes:
[0098] Ant colony selection unit S501, which is mainly used to select target ant colonies.
[0099] The initialization unit S502 is mainly used to initialize the parameters of the ant colony algorithm and the protein conformation, and establish the mapping relationship between the protein side chain conformation and the side chain rotamer library.
[0100] For the initialization unit 502, when the ant colony selection unit 501 selects p target ant colonies, the initialization unit
[0101] 502 When initializing the parameters of the ant colony algorithm, p ant colonies can share one pheromone matrix.
[0102] The main function of the rotame...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com