Biomarkers for HEDGEHOG inhibitor therapy
A technology of biomarkers and inhibitors, applied in the field of personalized therapy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] The multigene Hedgehog signature was originally developed by analyzing 40FFPE medulloblastoma (MB) samples. Matched fresh-frozen samples from the same cases were profiled previously as described by Y-J Cho and colleagues (Y-J Cho et al., JCO, 2010), and each individual case was identified as Hedgehog activated or non-Hedgehog activated based on its gene expression profile. A set of 32 candidate genes differentially expressed in hedgehog-positive (Hh+) versus hedgehog-negative (Hh-) tumors were selected from a comprehensive database of available profiling studies and an additional 22 potential normalizing genes. All candidate genes showing differential Hh+ versus Hh- expression were consistent across all profiling studies evaluated. RT-PCR assays for these candidate genes were developed and optimized for use with FFPE samples. Candidate genes that do not show robust expression within FFPE sample types are removed in further applications. In terms of 10 up- and 8 down-r...
Embodiment 2
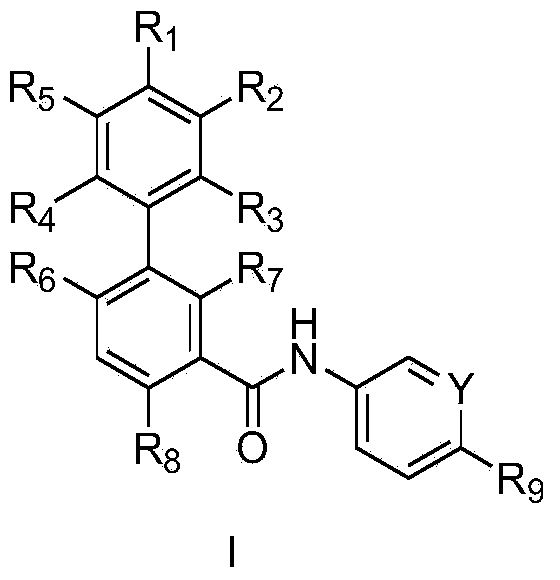
[0106] Recruitment of adult cancer patients, including medulloblastoma, into an ongoing study of methyl-4'-trifluoromethoxy-biphenyl-3-carboxylic acid [6-(cis-2,6-dimethyl -Morpholin-4-yl)-pyridin-3-yl]-amide Phase I dose escalation trial to evaluate methyl-4'-trifluoromethoxy-biphenyl-3-carboxylic acid [6-(cis -2,6-Dimethyl-morpholin-4-yl)-pyridin-3-yl]-amide safety and tolerability and evaluation of methyl-4'-trifluoromethoxy in adult patients - Maximum tolerated dose of biphenyl-3-carboxylic acid [6-(cis-2,6-dimethyl-morpholin-4-yl)-pyridin-3-yl]-amide. Archived FFPE tumor samples collected prior to the start of the trial were available for analysis by the method of the present invention from 3 medulloblastoma patients recruited into this trial. A daily dose of 200 mg methyl-4'-trifluoromethoxy-biphenyl-3-carboxylic acid [6-(cis-2,6-dimethyl-morpholin-4-yl)-pyridine-3- Base]-amide-treated patients achieved a RECIST-criteria partial response (PR) after 2 months of treatmen...
Embodiment 3
[0109] The method of the present invention was used to confirm Hedgehog activation status on another group of 40 medulloblastomas with a pathologically confirmed diagnosis. Of the 40 cases, 26 had classic medulloblastoma histology, 12 had nodular / desmoplastic histology, and 1 had large cell / undifferentiated histology. No other demographic data related to these cases were available. The Hedgehog activation status of these tumors was previously characterized by the affymetric gene expression profiling method as described by Y-J Cho and colleagues (YJ Cho et al. JCO, 2010). Hedgehog activation in 15 cases and non-Hedgehog activation in the remaining 25 cases were determined by the method of the present invention according to the 8-gene model, 5-gene model and each single gene SPHK1, OTX2, SFRP1, PDLIM3 or SHROOM2 in FFPE tumor samples Confirm in .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com