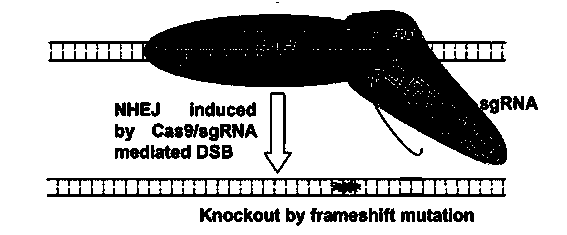
Method for human PD1 gene specific knockout through CRISPR-Cas9 (clustered regularly interspaced short palindromic repeat) and sgRNA(single guide RNA)for specially targeting PD1 gene
A pgl3-u6-hpd1sg, specific technology, applied in the field of genetic engineering, can solve problems such as laborious, time-consuming and expensive antibody drugs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 Design and synthesis of sgRNA for specifically targeting PD1 gene in CRISPR-Cas9 specific knockout of human PD1 gene
[0061] 1. Design of sgRNA targeting human PD1 gene:
[0062] (1) Select the 5'-GGN(19)GG sequence on the PD1 gene. If there is no 5'-GGN(19)GG sequence, 5'-GN(20)GG or 5'-N(21)GG is also Can.
[0063] (2) The target site of sgRNA on the PD1 gene is located in the exon of the gene, which is more likely to cause the deletion of the fragment or the frame-shift mutation, so as to achieve the purpose of complete gene inactivation.
[0064] (3) The targeting site of sgRNA on the PD1 gene is located on the common exons of different splicing forms.
[0065] (4) Use BLAT in the UCSC database or BLAST in the NCBI database to determine whether the target sequence of the sgRNA is unique and reduce potential off-target sites.
[0066] According to the above method, we designed a total of 180 sgRNAs targeting the human PD1 gene, the sequences of which ar...
Embodiment 2
[0087] Example 2 Using CRISPR-Cas9 to specifically knock out the human PD1 gene (the sgRNA used to target the PD1 gene is shown in Sequence Table 52)
[0088] 1. The linearized sequence is the pGL3-U6-sgRNA plasmid shown in the sequence table SEQ ID NO.10.
[0089] Enzyme digestion system and conditions are as follows:
[0090] 2 μg pGL3-U6-sgRNA (400ng / μl);
[0091] 1μl CutSmart Buffer;
[0092] 1 μl BsaI (NEB, R0535L);
[0093] Replenish water to 50 μl, incubate at 37°C for 3-4 hours, shake and centrifuge at regular intervals to prevent droplets from evaporating onto the tube cap.
[0094] After digestion, use AxyPrep PCR Clean up Kit (AP-PCR-250) to purify and recover to 20-40 μl sterilized water.
[0095] 2. Combine the double-stranded sgRNA oligonucleotides obtained after denaturation and annealing that can be connected to the U6 eukaryotic expression vector (the sequences of their Forward oligo and Reverse oligo are shown in the sequence table SEQ ID NO.1 and 2, resp...
Embodiment 3
[0125] Example 3 Using CRISPR-Cas9 to specifically knock out the human PD1 gene (the sgRNA used to target the PD1 gene is shown in the sequence table as SEQ ID NO.82)
[0126] 1. The linearized sequence is the pGL3-U6-sgRNA plasmid shown in the sequence table SEQ ID NO.10.
[0127] Enzyme digestion system and conditions are as follows:
[0128] 2 μg pGL3-U6-sgRNA (400ng / μl);
[0129] 1μl CutSmart Buffer;
[0130] 1 μl BsaI (NEB, R0535L);
[0131] Replenish water to 50 μl, incubate at 37°C for 3-4 hours, shake and centrifuge at regular intervals to prevent droplets from evaporating onto the tube cap.
[0132] After digestion, use AxyPrep PCR Clean up Kit (AP-PCR-250) to purify and recover to 20-40 μl sterilized water.
[0133] 2. The double-stranded sgRNA oligonucleotides obtained after denaturation and annealing that can be connected to the U6 eukaryotic expression vector (the sequences of their Forward oligo and Reverse oligo are shown in the sequence table SEQ ID NO.3 an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com