Host cells and methods for production of isobutanol
A technology of host cells and recombinant host cells, which is applied in the field of recombinant host cells and can solve problems such as high price and non-environmental protection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
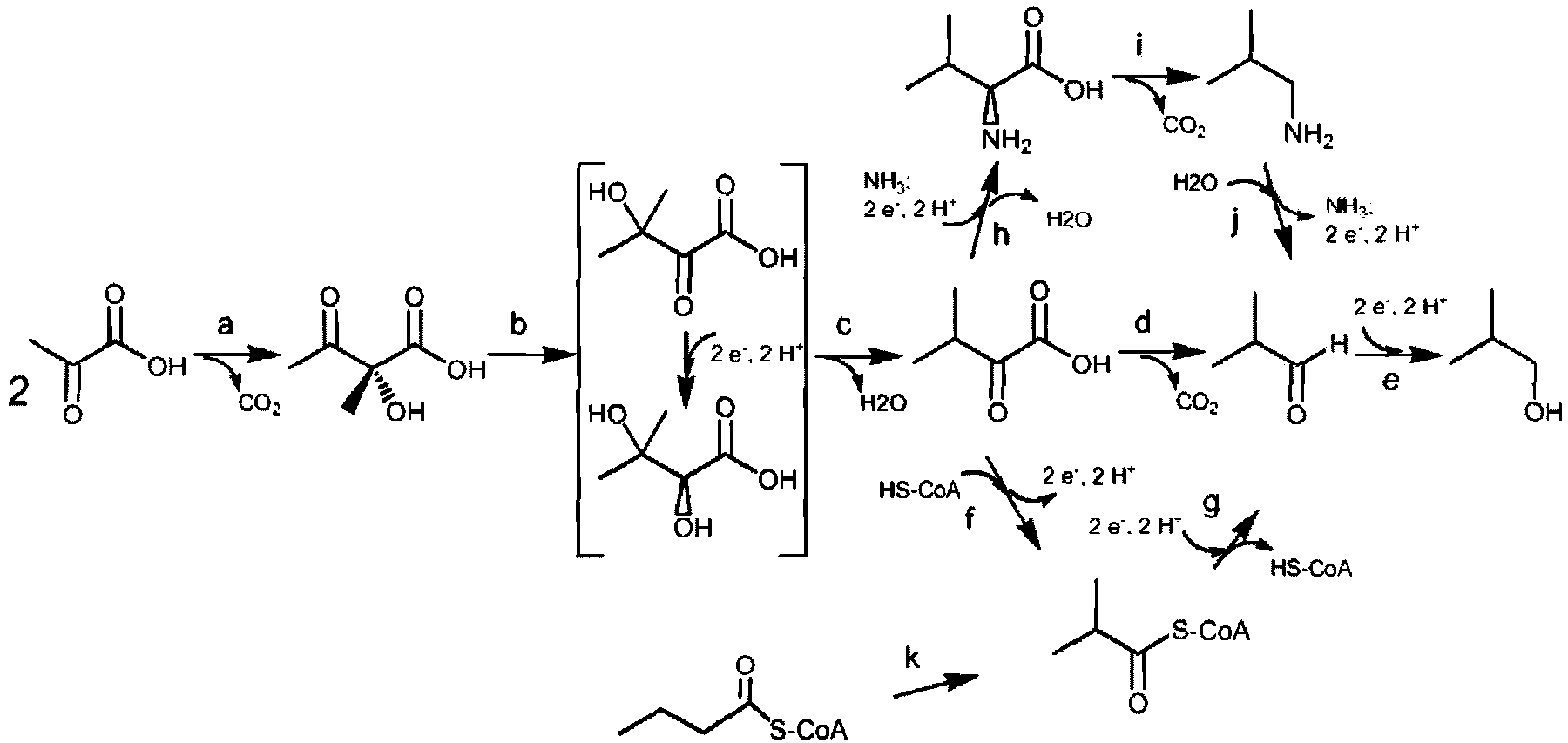
[0208] A biodiversity screen for KARI-encoding genes from various bacterial and fungal species is described in Example 1, showing suitable KARIs for isobutanol production. Using the present invention, those skilled in the art should readily be able to identify other suitable polypeptides having KARI activity.
[0209] Sequences of other polynucleotides, genes and / or polypeptides can be identified from the literature and from bioinformatics databases well known to those of skill using the sequences disclosed herein and available in the art. For example, such sequences can be identified by BLAST searches of publicly available databases with the polynucleotide or polypeptide sequences provided herein. In such methods, the identity can be based on the Clustal W alignment method using the default parameters of GAP PENALTY=10, GAP LENGTHPENALTY=0.1 and a protein weight matrix of the Gonnet 250 series.
[0210] In addition, the polynucleotide or polypeptide sequences disclosed herein ...
example
[0460] The invention will be further defined in the following examples. It should be understood that these Examples, while indicating preferred embodiments of the invention, are given by way of illustration only. Through the above discussion and these examples, those skilled in the art can ascertain the essential characteristics of the present invention, and without departing from the spirit and scope of the present invention, various changes and modifications can be made to the present invention to adapt to various applications and conditions .
[0461] general method :
[0462] Standard recombinant DNA and molecular cloning techniques used in the examples are well known in the art and described in: Sambrook, J., Fritsch, E.F. and Maniatis, T., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989, T.J. Silhavy, M.L; Bennan and L.W. Enquist, Experiments with Gene Fusions, Cold Spring Harbor Laboratory, Cold Spring Harbor...
example 2
[0490] Screening of the KARI diversity collection for isobutanol production
[0491] The various KARI genes were evaluated based on their "effective yield" in yeast. The effective yield is measured after a certain period of growth (for example, 48 hours) under progressive oxygen-limited conditions. Yeast biomass at 1OD 600 The yeast cells are equivalent to 0.3g / L to calculate the assumption.
[0492] Yeast isobutanol pathway strains carrying different KARI genes were inoculated into 10 ml of SEG-Ura, His medium with 0.2% glucose and 0.2% ethanol and allowed to grow aerobically at 30°C overnight to about 2OD. The culture was centrifuged and a portion of the cells resuspended in SEG-Ura, His (2% glucose, 1% ethanol) in a 125ml shake flask to an initial OD of 0.4 in a total volume of 25ml 600 . The shake flasks were closed with screw-on solid plastic caps and cultures were grown in the flasks under progressively oxygen-limited conditions with minimal air and oxygen exchange...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com