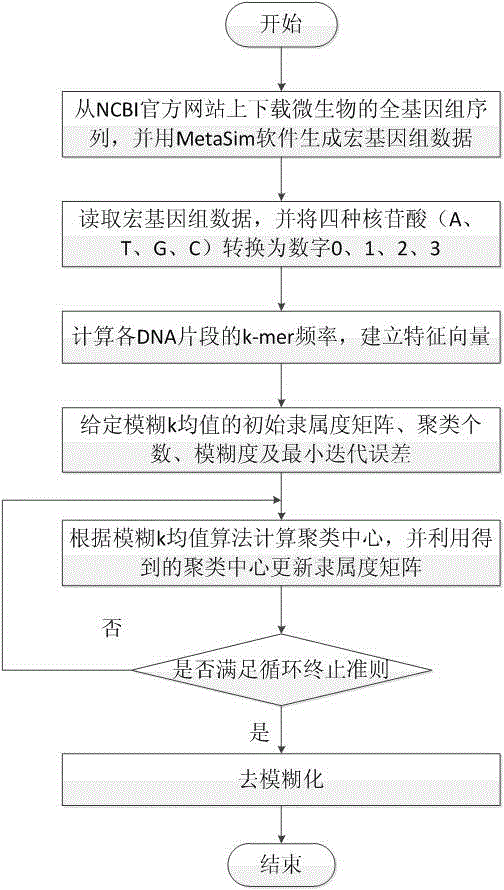
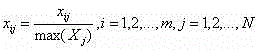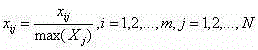Micro genome segment clustering method based on fuzzy k-mean
A technology of metagenomics and clustering methods, which is applied in the fields of instruments, calculations, electrical digital data processing, etc., and can solve problems such as errors, misclassifications, and easy pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0099] Example: Cluster analysis of simulated metagenomic data
[0100] The metagenomic data in this embodiment is simulated from the whole genome data of 20 species, which contains 20 species, each species has 300 DNA fragments, and the length of each fragment is 200nt (gene fragment length unit, meaning nucleotides), therefore, there are a total of 6000 DNA fragments in the dataset. We use the method proposed by the present invention to perform feature extraction, data normalization, and cluster analysis on the metagenomic data in this example. The metagenomic data information in the present embodiment is as shown in Table 1, and the result information of cluster analysis is as shown in Table 2 using the method proposed by the present invention:
[0101] Table 1: Metagenome data information in this embodiment
[0102]
[0103] Table 2: Results of Cluster Analysis
[0104]
[0105] It can be seen from Table 2 that the cluster analysis of the 20 species in this examp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com