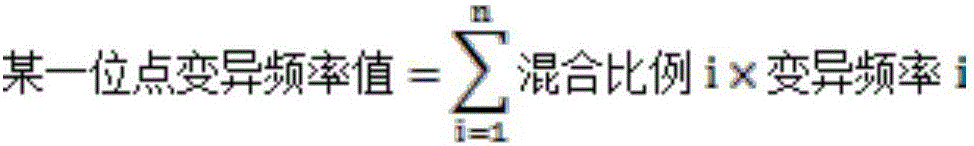Quality control method for detecting human EGFR (Epidermal Growth Factor Receptor) gene variation based on high-throughput sequencing and kit
A gene mutation and kit technology, which is used in the determination/inspection of microorganisms, biochemical equipment and methods, DNA preparation, etc., to achieve the effect of improving efficiency and reducing the cost of detection quality control
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Preparation of Quality Control Products for Human Circulating Tumor DNA EGFR Gene Variation Detection
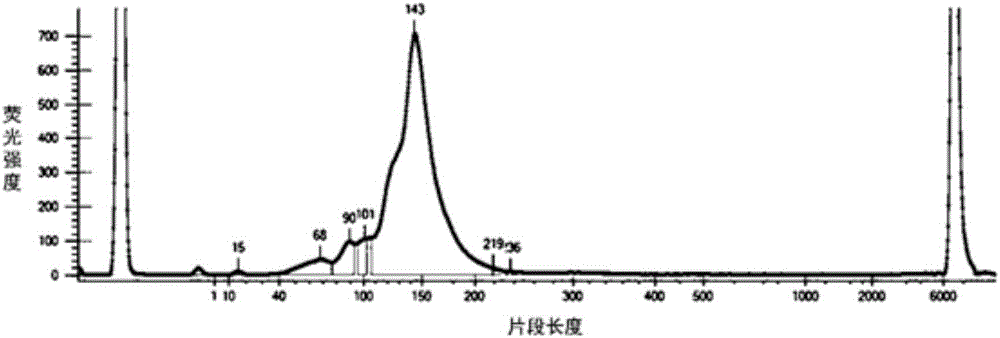
[0040] 1.1. Six commonly used stable human tumor cell lines A549, NCI-H720, NCI-H1650, NCI-H1975, SW48 and SW1417 were purchased from ATCC. The genomic DNA of each cell line was sequenced by the Sanger method, and the heterozygous and homozygous mutation sites confirmed by the Sanger method were used as positive control sites. It has been verified that there are 12 positive mutation sites in total.
[0041] 1.2. These six human tumor cell lines were cultured in a special medium, culture conditions: 37°C constant temperature, 5% CO2, humidity 50%. Cultivate until the cell density reaches 80-90% of the area of the culture dish, collect the cells in the logarithmic phase of growth, centrifuge at 1500g at 4°C for 5min, take the precipitate and discard the supernatant.
[0042] 1.3. Resuspend in pre-cooled PBS solution, centrifuge at 5000rpm at 4°C for 5min,...
Embodiment 2
[0046] Example 2: Preparation of Human Circulating Tumor DNA EGFR Gene Variation Detection Kit
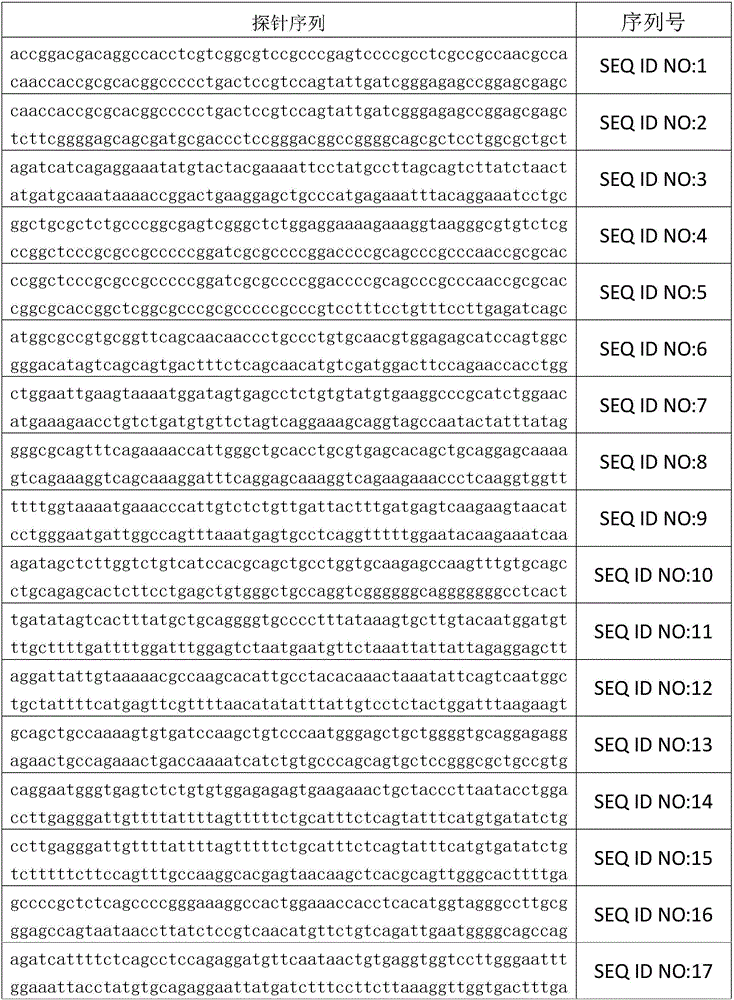
[0047] 2.1. Design and synthesize multiple capture probes for different target regions on the human circulating tumor DNA EGFR gene. The collection of all capture probes can cover all coding exon regions and exon-intron junction regions of the human EGFR gene; The capture probe is labeled with biotin; the sequence of the capture probe is shown in SEQ ID NO: 1-195 below.
[0048]
[0049]
[0050]
[0051]
[0052]
[0053]
[0054]
[0055]
[0056]
[0057]
[0058]
[0059] 2.2. Mix all the capture probes represented by SEQ ID NO: 1-195 in the above table in the same ratio, and dilute the mixture to a working concentration of 1.5 PM (PM = picomole / liter), and store at -20°C.
[0060] 2.3. Separately pack the capture probe mixture and the quality control obtained in Example 1.
[0061] 2.4. Prepare instructions, outer packaging, assemble and seal....
Embodiment 3
[0063] Example 3: Sequencing detection of human circulating tumor DNA EGFR gene variation
[0064] The instrument used for sequencing in this example is the gene sequence automatic analyzer CN500.
[0065] The preparation method of the quality control product is the same as that in Example 1.
[0066] 3.1. Human cfDNA was extracted from 15 positive plasma samples, and quality control products were used directly without extraction.
[0067] 3.2. Library construction
[0068] 3.2.1. End Repair
[0069] Add 30ng of extracted cfDNA sample or quality control to 0.2ml PCR reaction tube, make up to 50μL with Low EDTA TE, vortex to mix, and centrifuge briefly. Add 1 μL of end-repair enzyme mixture and 6 μL of end-repair reaction buffer, vortex and mix briefly, and then incubate at 37°C for 5 minutes, then incubate at 65°C for 30 minutes, and then at 37°C for 5 minutes for end-repair. After the reaction, take out the PCR reaction tube and transfer all the products to a new 1.5mL ce...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com