Polynucleotides, polypeptides and methods for enhancing photossimilation in plants
A polynucleotide and plant technology, applied in the field of molecular biology, can solve labor-intensive and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0129] Example 1: Identifying Candidates
[0130] This example describes a genetic engineering strategy to enhance photoassimilation in maize and other NADP malate C4 species. One computer model output organized into combined solutions for 3 and 4 genes. A 3-gene and a 4-gene combination were each selected for trait development. To implement this trait, query the BRENDA database (brenda.enzymes.org) for information on phosphoenolpyruvate carboxylase (PEPC, EC 4.1.1.31), fructose-1,6-bisphosphate Enzyme (FBPase, EC3.1.3.11), phosphoribulokinase (PRK, EC2.7.1.19), NADP-malate dehydrogenase (NADPME, EC1.1.1.82) and pyruvate orthophosphate dikinase ( Sequence information of PPDK, EC2.7.9.1). This analysis provides the protein sequence of a functionally characterized enzyme. Information from this database was used to obtain the protein sequences of PEPC from maize, FBPase from spinach, phosphoribulokinase from spinach, and NADP-malate dehydrogenase from sorghum. Briefly, refer...
example 2
[0131] Example 2: Regulatory Sequences Targeting Expression of Candidate Genes
[0132] Once candidate genes have been identified, regulatory sequences are selected to target the expression of these candidate genes to appropriate cell types. Design of a series of plant expression cassettes to deliver robust trait gene expression in mesophyll or bundle sheath cells. A combination of proteomic data (Majeran, W. et al. (2005) Plant Cell, 17:3111-3140) and expression profiling data was used to identify candidate regulators based on the expression pattern of the gene of interest. sequence, and identified six new expression cassettes (Coneva V, et al. (2007) J of Exp Botany 58:3679-3693). Each cassette consists of a promoter and terminator sequence. The promoter consists of a 5'-untranscribed sequence (the first intron) and a 5'-untranslated sequence (consisting of the first exon and part of the second exon). In addition, the promoter is derived from the tobacco mosaic virus sequ...
example 3
[0136] Example 3: Expression Cassettes and Combinations
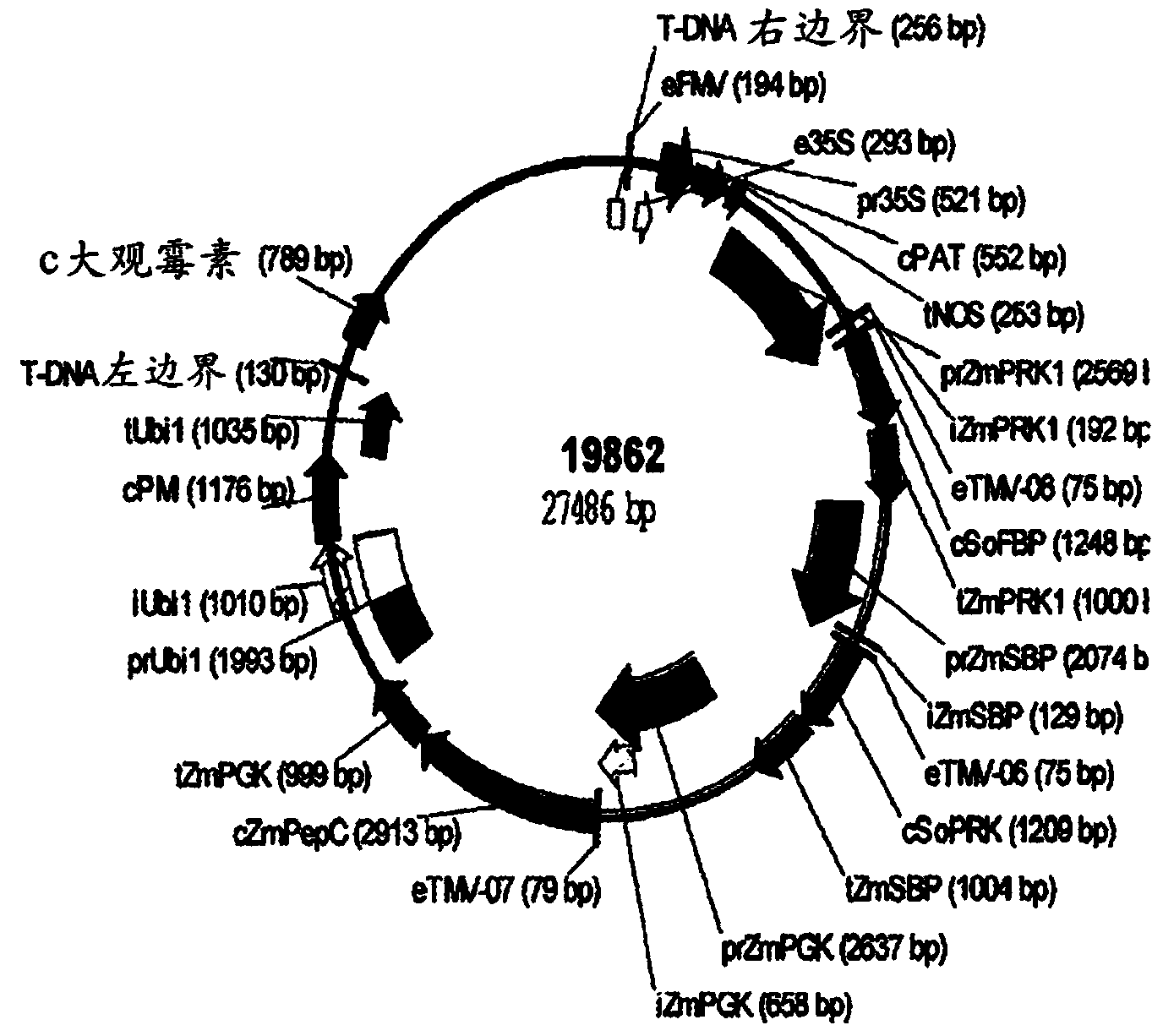
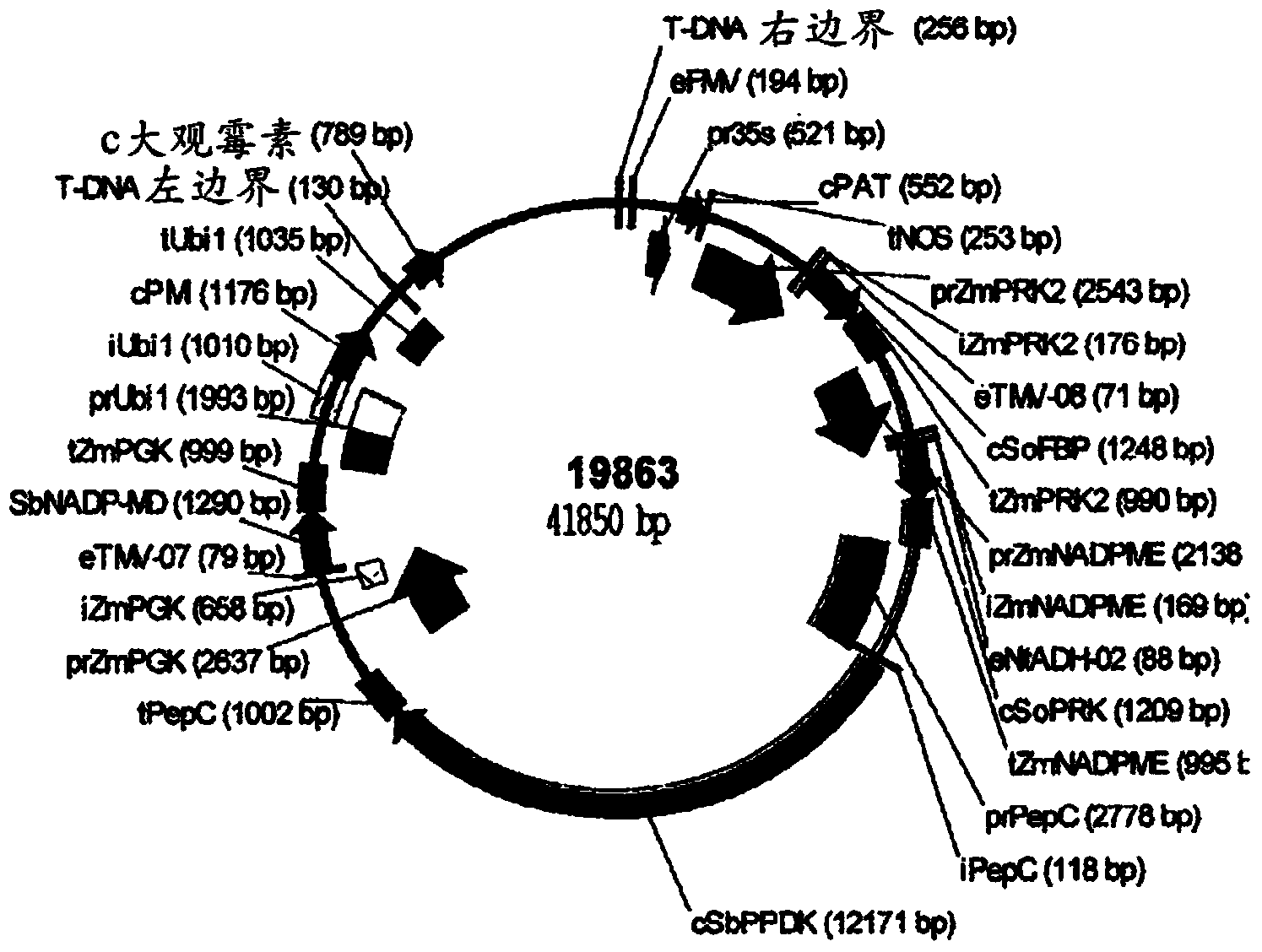
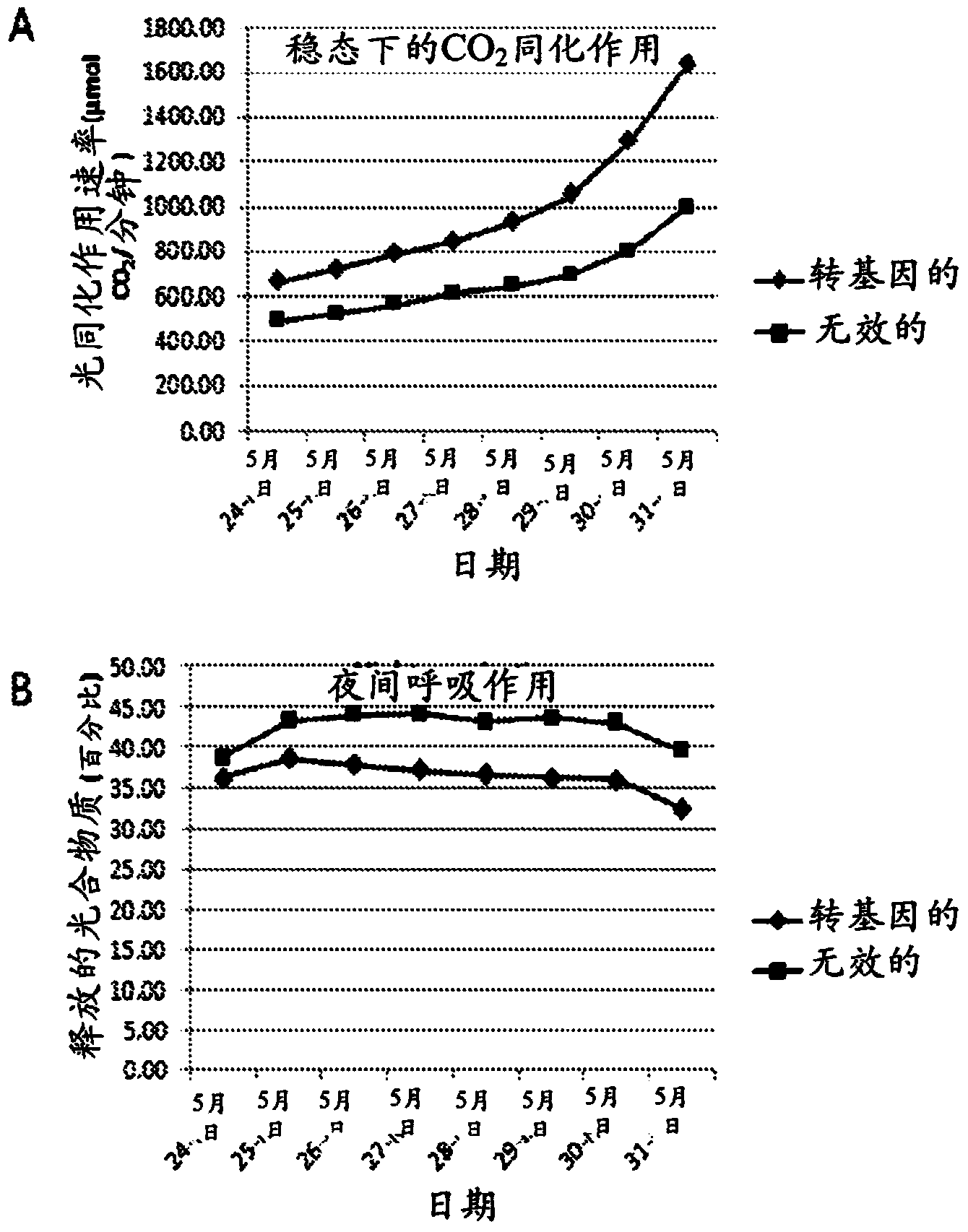
[0137] One three-gene and one four-gene expression cassette binary vectors containing candidate genes selected by the method of the present invention will each be used to reduce C4 photosynthetic model output to practice. The C4 photosynthesis enhancing constructs of the three genes are shown in Table 2; the C4 photosynthesis enhancing constructs of the four genes are shown in Table 3. Gene numbers indicate the sequence, starting at the right border of the T-DNA and extending to the left border. The binary vector for this trigene is 19862 and is shown in figure 1 middle. The binary vector for the four genes is 19863 and is shown in figure 2 middle.
[0138] Table 2.
[0139]
[0140] table 3.
[0141]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com