FRM1 gene CGG sequence repeat detection method and application
A detection method and gene sequence technology, applied in the fields of biotechnology and medicine, can solve the problems of difficult extension of FMR1 gene DNA unzipping primers, high incidence of Fragile X syndrome, complex clinical manifestations, etc., and achieve a wide range of amplification conditions. , moderate length, simple use of instruments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0040] The present invention will be further described below in conjunction with the accompanying drawings and examples, but not as a limitation to the present invention.
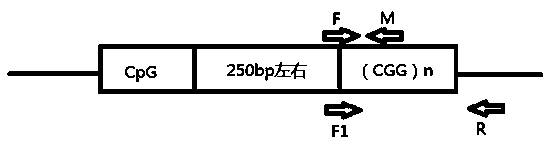
[0041] like figure 1 As shown, in order to solve the above problems, the present invention designs a pair of internal reference primers: forward primer F and reverse primer M, and the size of the amplified fragment is about 220bp, which is used to verify that the PCR reaction is feasible and calculates the number of CGG repeats benchmark.
[0042] Set a pair of detection primers downstream of the CGG sequence repeat region of FMR1 gene, forward primer F1 and reverse primer R, the size of the amplified fragment is about (470+3n) bp (n represents the number of CGG repeats), used to determine the CGG sequence repeats The area where the number is located.
[0043] SEQ NO.1 F: 5'-CGACCTGTCACCGCCCTTCAGCCTTCC-3'
[0044] SEQ NO.2 M: 5'-CGCTGCGGGTGTAAACACTGAAACCACGTC-3'
[0045] SEQ NO.3 F1: 5'-CGACCTGTCACCGCC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com