Identification of multigene biomarkers
A gene and population technology, applied in the direction of microbial measurement/testing, biochemical equipment and methods, etc., can solve the problems of patients and restrictions that cannot choose molecularly defined cancers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0168] Example 1: Murine Tumor-BH Archive
[0169] A genetically diverse population of more than 100 murine mammary tumors (BH archive) was used to identify tumors sensitive to a drug of interest (responders) and tumors resistant to the same drug of interest (non-responders). The BH archive was established by in vivo propagation and cryopreservation of primary tumor material from more than 100 spontaneous murine mammary tumors derived from engineered chimeric mice, and the Mice develop HER2-dependent, inducible spontaneous mammary tumors.
[0170] Mice were formed basically as follows. Ink4a homozygous nude mouse ES cells were co-transfected with the following four constructs (as isolated fragments): MMTV-rtTA, TetO-HER2 V659Eneu , TetO-luciferase and PGK-puromycin. ES cells carrying these constructs were injected into 3-day-old C57BL / 6 embryo sacs, and the embryo sacs were transplanted into pseudopregnant female mice to make them pregnant and give birth to chimeric mice. ...
Embodiment 2
[0174] Example 2: Identification of Tivozanib PGS
[0175] Tumors in BH mouse tumor archives were assayed for sensitivity to treatment with tivozanib. Evaluation of the response of the tumor to the drug treatment was performed basically as follows. Subcutaneously implanted tumors were established by injecting physically disrupted tumor cells (mixed with Matrigel) into 6-week-old female BALB / c nude mice. When the tumor reached about 100-200mm3, 20 tumor-bearing mice were randomly divided into 2 groups. Group 1 received vehicle. Group 2 received tivozanib at 5 mg / kg daily by oral gavage. Tumors were measured twice a week using calipers, and tumor volumes were calculated.
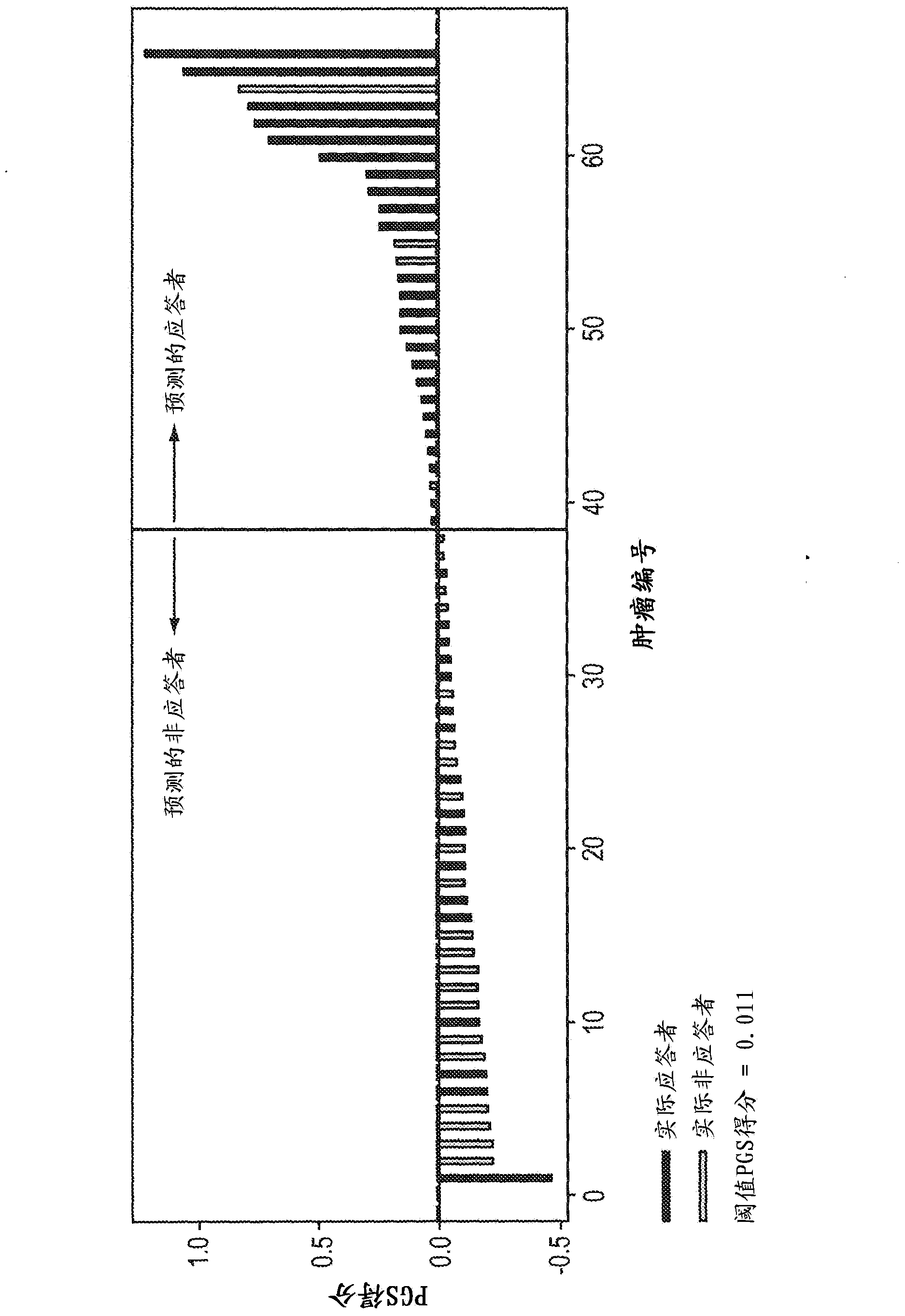
[0176] These studies demonstrate significant tumor-to-tumor variability in growth inhibition in response to tivozanib. This change in response was expected because mouse model tumors propagated from spontaneously arising tumors and were therefore expected to include a different set of secondary de novo mu...
Embodiment 3
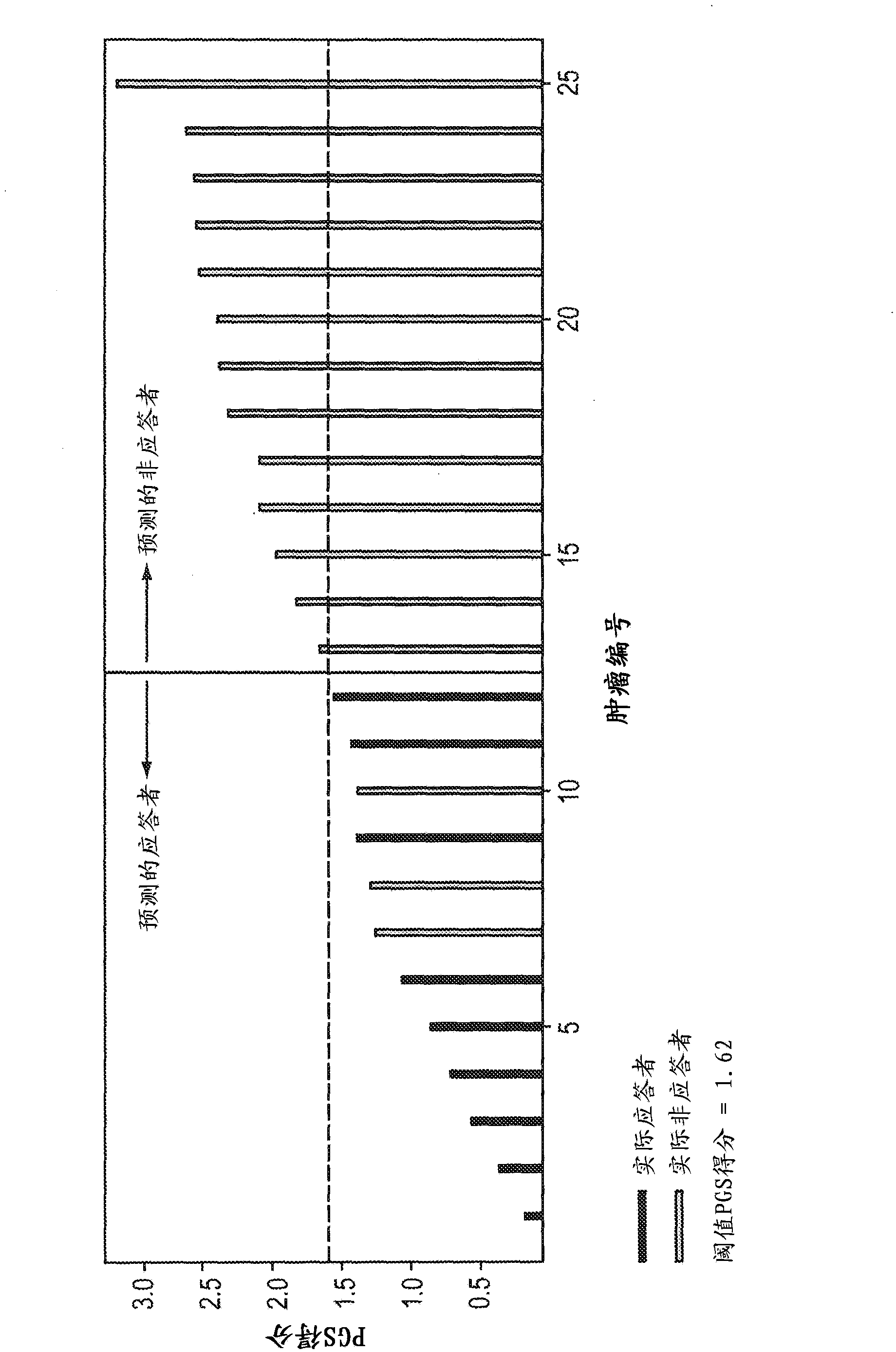
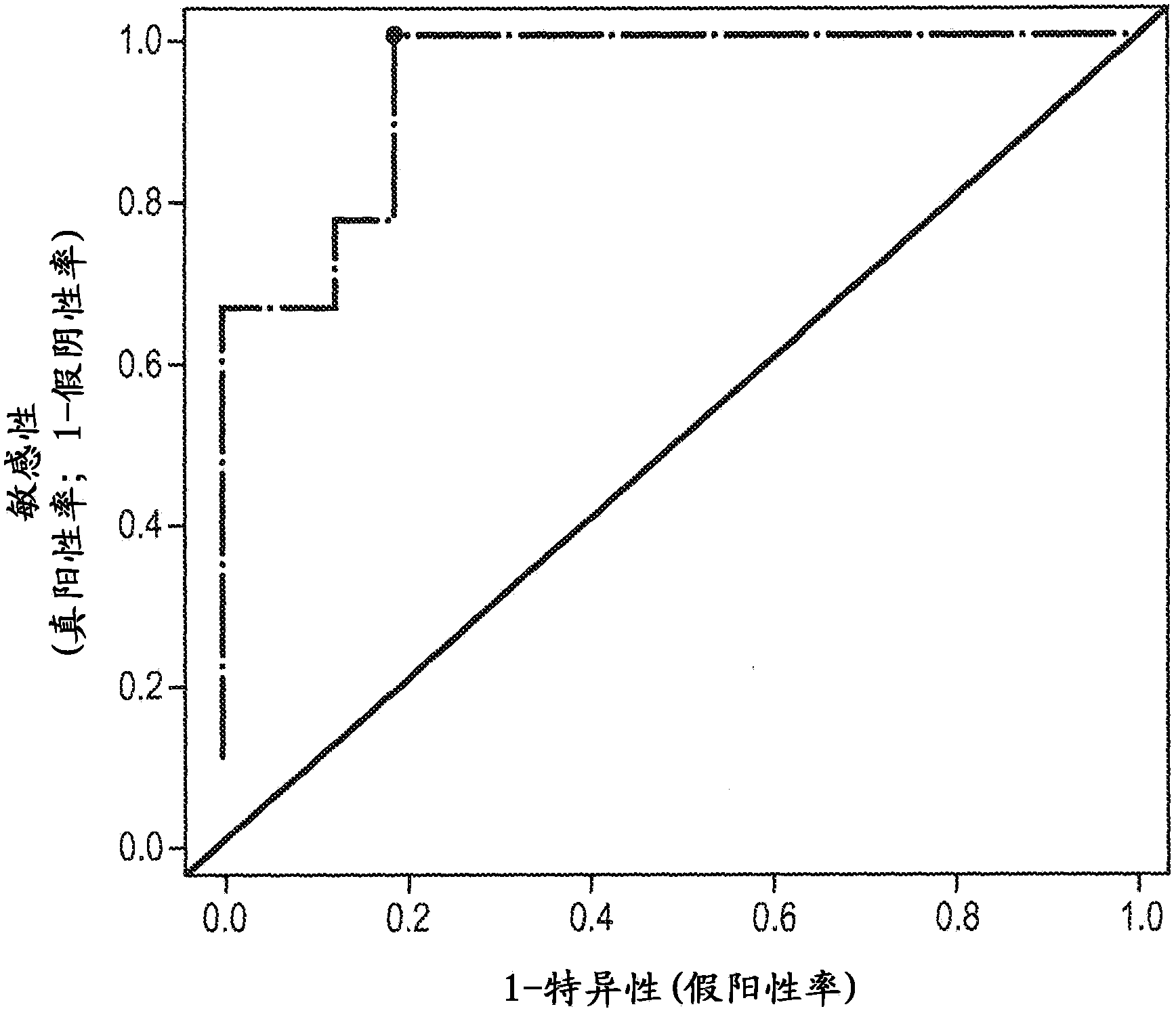
[0184] Example 3: Predicting mouse response to tivozanib
[0185] The predictive power of the tivozanib PGS (TC50) identified in Example 2 was evaluated in a trial involving 25 tumor populations previously based on actual experience with tivozanib as described in Examples 1 and 2. Drug response assays were classified as tivozanib-sensitive or tivozanib-resistant. These 25 tumors were obtained from a proprietary archive of primary mouse tumors in which the driver proto-oncogene was HER2. In this example, the PGS used was the following 10-gene subset derived from TC50:
[0186] MRC1
[0187] ALOX5AP
[0188] TM6SF1
[0189] CTSB
[0190] FCGR2B
[0191] TBXAS1
[0192] MS4A4A
[0193] MSR1
[0194] NCKAP1L
[0195] FLI1
[0196] The PGS score for each tumor was calculated from the gene expression data obtained by conventional microarray analysis. We calculated the tivozanib PGS score according to the following algorithm:
[0197]
[0198] Among them, E1, E2...En ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com