Molecular marker for discriminating different ecotype populations of coilia ectenes
A molecular marker and eco-type technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of unseen research reports and the scarcity of SINE molecular markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Experimental Materials
[0025] According to the source of the samples, the anchovy is divided into 6 populations: Xiangshan Port (XS) in Zhejiang, Chongming in Shanghai (CM), Jingjiang in Jiangsu (JJ), Lake Taihu (TH), Poyang Lake in Jiangxi (PY), and Dongting Lake in Hunan (DT). The sample size of each population is 10 tails. The samples from Xiangshan Port in Zhejiang came from the aquatic product market, and the collection place is unknown, and the rest of the samples were caught on board.
[0026] Genomic DNA Extraction
[0027] The extraction of genomic DNA from the sample was performed according to the instructions of Shanghai Sangon’s UNIQ-10 Column Genomic DNA Extraction Kit. The quality and concentration of the extracted DNA were detected by an ultraviolet spectrophotometer, and stored at -20°C for future use.
[0028] site segregation
[0029] Primers were designed according to the conserved sequence of SINE, and the partial sequence of SINE in the a...
Embodiment 2
[0048] experimental method
[0049] The specific primer of SINE insertion site, its sequence is:
[0050] The upstream primer at the Locus 18 site is shown in SEQ ID NO.1,
[0051] The downstream primer at the Locus 18 site is shown in SEQ ID NO.2,
[0052] The upstream primer of the Locus 40 site is shown in SEQ ID NO.3,
[0053] The downstream primer at the Locus 40 site is shown in SEQ ID NO.4,
[0054] The upstream primer at the Locus 58 site is shown in SEQ ID NO.5,
[0055] The downstream primer at the Locus 58 site is shown in SEQ ID NO.6.
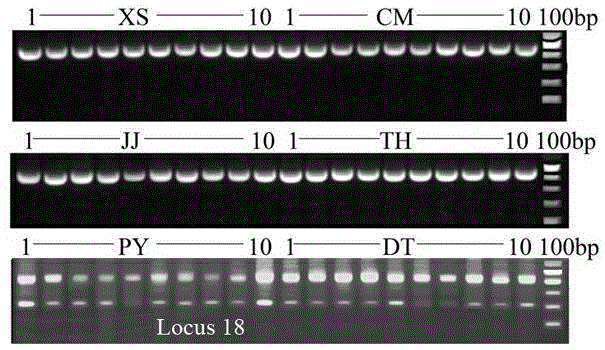
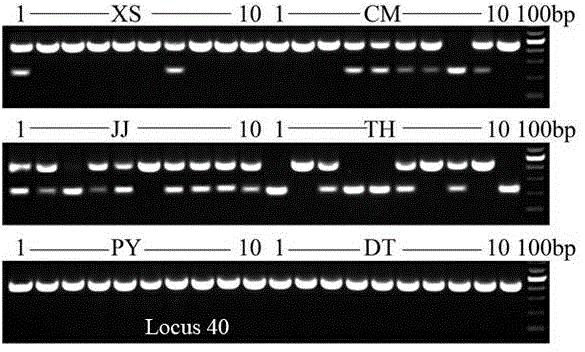
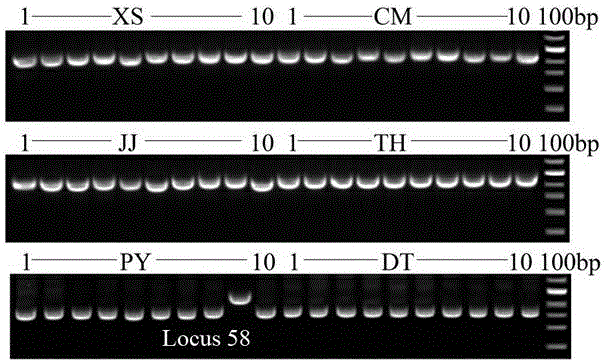
[0056] Through PCR amplification, agarose gel electrophoresis, cloning, and sequencing, the population-specific DNA fragments of the SINE locus were obtained, and different populations of the anchovy were identified according to the differences in the number and size of the amplified fragments.
Embodiment 3
[0058] experimental method
[0059] The specific primer of SINE insertion site, its sequence is:
[0060] The upstream primer at the Locus 18 site is shown in SEQ ID NO.1,
[0061] The downstream primer of the Locus 18 site is shown in SEQ ID NO.2.
[0062] Through PCR amplification, agarose gel electrophoresis, cloning, and sequencing, the population-specific DNA fragments of the SINE locus were obtained, and the sequencing results of the Locus 18 locus were analyzed, and a band with a size of about 450 bp could be amplified but not The samples with 200bp amplified bands are the anchovy from Xiangshan Port, Chongming, Jingjiang or Taihu Lake, and the samples that can amplify 450 and 200bp bands are the anchovy from Poyang Lake or Dongting Lake.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com