hmg1 Gene and Its Application in Molecular Detection of Microsporidium Bombyx mori
A technology of microsporidia and silkworm, which is applied in application, genetic engineering, plant genetic improvement, etc., can solve the problems of low primer sensitivity, low sensitivity, detection interference, etc., and achieve the effects of wide application range, convenient use, and increased range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 The HMG1 gene of the silkworm Microsporidia
[0040] 1. According to the method of gene homologous cloning in the gene cloning technology of molecular biology, the full-length cDNA and DNA sequences of the HMG 1 gene of N. silkworm were cloned.
[0041] 2. Obtaining the full length of cDNA, the specific method is as follows:
[0042] (1) Using Primer premier 5.0 software, combined with comprehensive analysis, the primers primers HMG1F / HMG1R were designed, and the sequences are shown in SEQ ID NO.3 and SEQ ID NO.4, respectively.
[0043] Upstream primer HMG1F (SEQ ID NO.3):
[0044] 5' ATGACTGCTCAAAAAGACGATAC 3'
[0045] Downstream primer HMG1R (SEQ ID NO.4):
[0046] 5'TTATTCATCACTATTCTCCTACTTCT 3'.
[0047] (2) Using the purified spore DNA of N.b. silkworm (N.b) as a template, PCR amplification was performed with primers HMG1F / HMG1R.
[0048] (3) The PCR product was purified, connected to pMD19T, and transformed into E, coli DH-5α for culture.
[0049] ...
Embodiment 2
[0053] Example 2 Detection primer design and establishment of PCR amplification method
[0054] 1. Primer design
[0055] On the basis of obtaining the HMG 1 gene of Bombyx mori, the Primer Premier 5.0 software was used to design multiple pairs of primers. After a large number of drug resistance, specificity and sensitivity tests, three pairs of primers were finally selected as representative primers. The primer sequences of each group are as follows:
[0056] (1) The first pair:
[0057] Upstream primer HMG1F (SEQ ID NO.3):
[0058] 5' ATGACTGCTCAAAAAGACGATAC 3'
[0059] Downstream primer HMG1R (SEQ ID NO.4):
[0060] 5'TTATTCATCACTATTCTCCTACTTCT 3'.
[0061] (2) The second pair:
[0062] Upstream primer HMG1-sF (SEQ ID NO.5):
[0063] TTCCGAAATAATCTTCTTTTAATTG
[0064] Downstream primer HMG1-sR (SEQ ID NO.6):
[0065] TTGTGCACCGAATCGTAAATAG
[0066] (3) The third pair:
[0067] Upstream primer HMG1-xF (SEQ ID NO.7):
[0068] TCCCTAGGAACTTTTAAAGAGAAG
[0069] Dow...
Embodiment 3
[0091] Example 3 Primer-specific detection
[0092] 1. The DNA of Microsporidia Bombyx (N.b), Microsporidia tussah (N.a), and Microsporidia corn borer (N.f) were used as templates, and the primers HMG1F / HMG1R, HMG1-sF / HMG1-sR, HMG1-xF / HMG1-xR, PCR amplification was carried out by the method of Example 2, and the results were detected by agarose gel electrophoresis after the amplification.
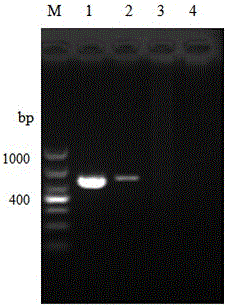
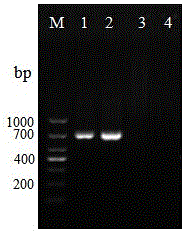
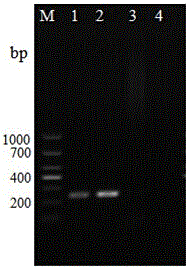
[0093] 2. The amplification results of the three pairs of primers are shown in the attached Figure 4~6 shown. The results showed that only the primer HMG1-sF / HMG1-sR could specifically detect Bombyx mori Microsporidia, while the primers HMG1F / HMG1R and HMG1-xF / HMG1-xR could detect all the microsporidia, and had good generality. Detectable, but not specific for Microsporidia Bombyx mori.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com