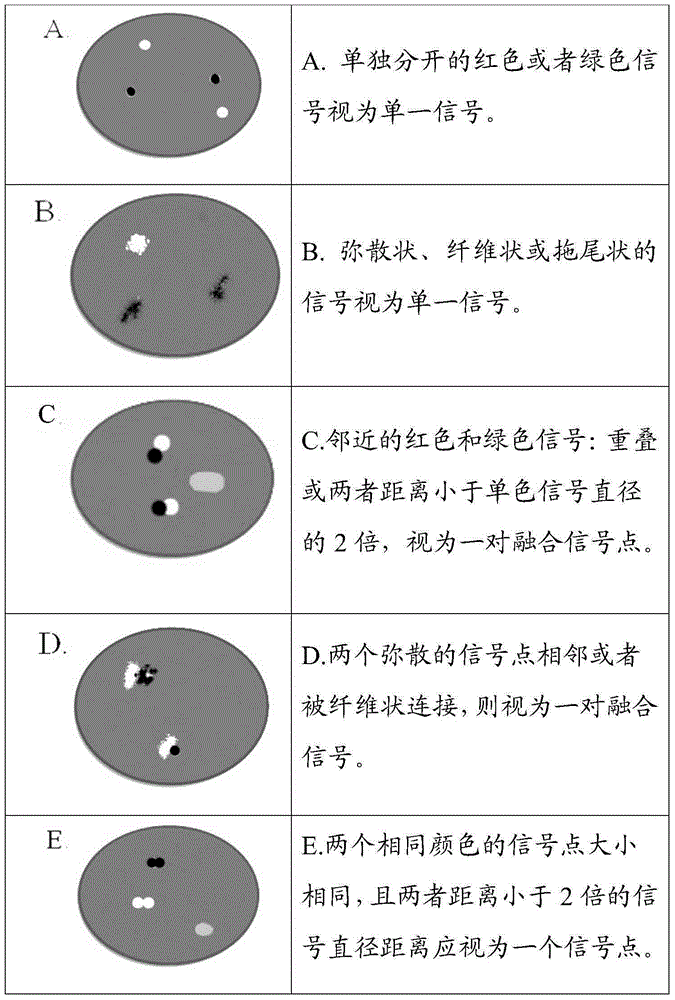
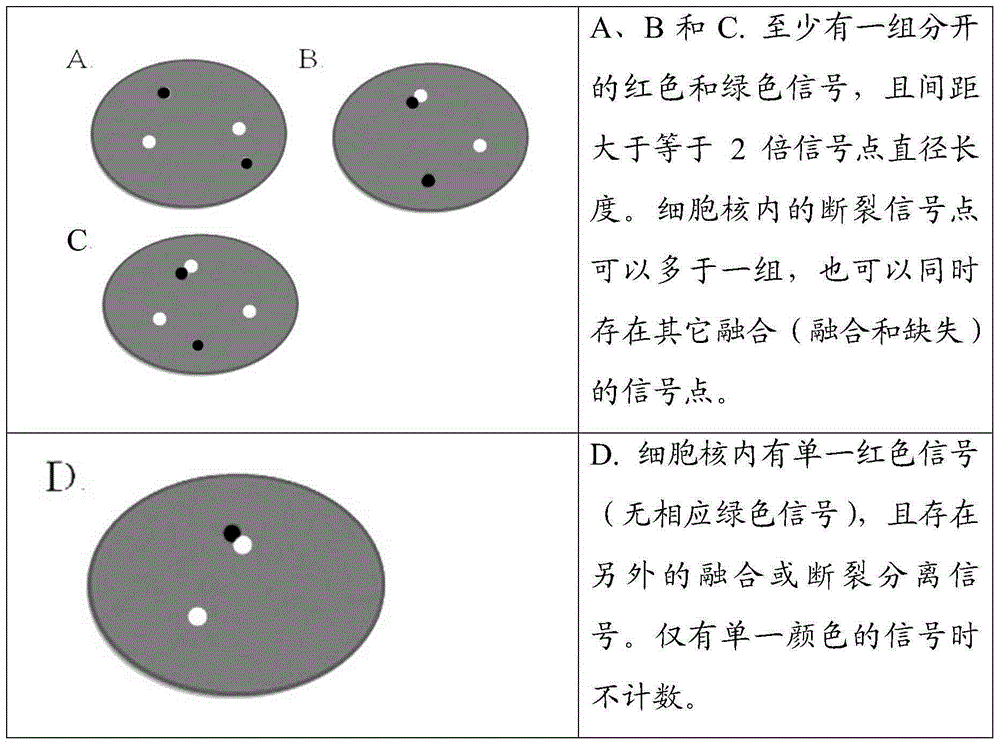
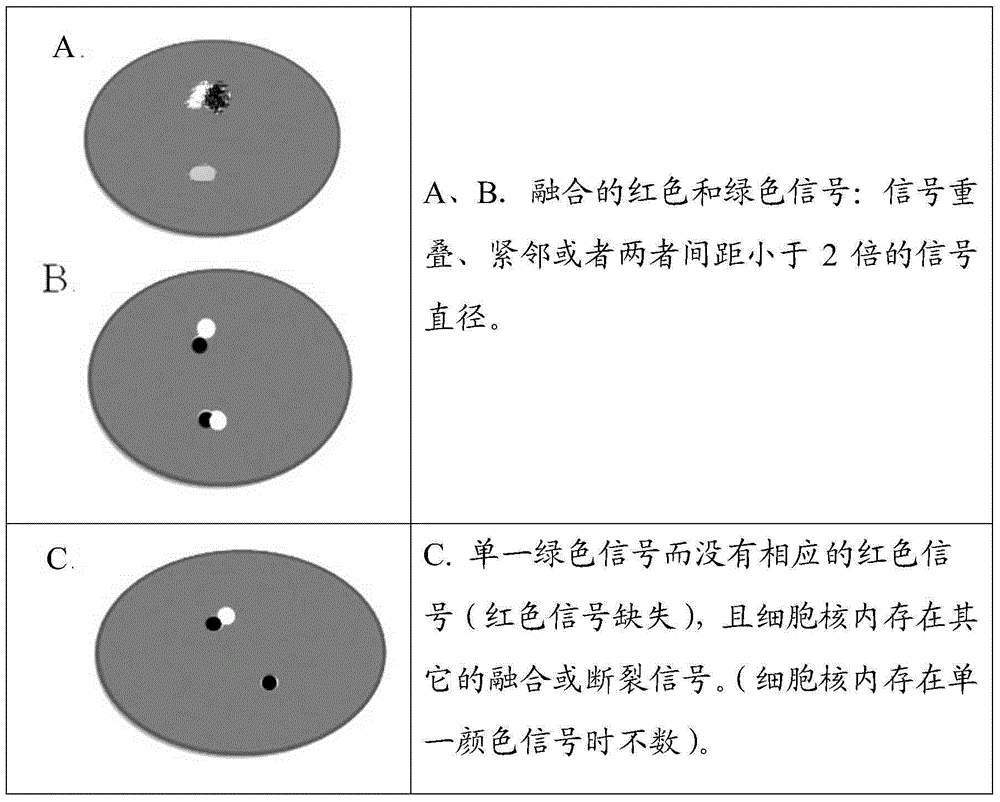
ALK gene rearrangement detection probe, kit and method
A detection kit and gene rearrangement technology, applied in the field of molecular biology, can solve the problems of inability to detect fusion type, reduce the specificity and sensitivity of FISH detection, and achieve the effects of fast detection speed, high accuracy and low false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The detection probe and kit for detecting ALK gene rearrangement described in this embodiment are designed as follows:
[0034] 1. Design the amplification primers
[0035] The ALK gene is located on chromosome 2 (position 2p23), and the breakpoint of the ALK gene rearrangement is located between exons 19 and 20. According to the characteristics of the ALK gene, two sets of amplification primers were designed respectively: for the breakpoint The first group of amplification primers designed upstream, the second group of amplification primers designed downstream of the breakpoint, and the corresponding amplification products respectively constitute the first group of probe libraries and the second group of probe libraries. For each set of amplification primers, the obtained amplification product can be used as a rearranged FISH probe, does not contain repeated sequences, can avoid cross-reaction, and can specifically recognize and hybridize with the ALK target detection ...
Embodiment 2
[0088] Example 2 Detection of clinical samples using the ALK gene rearrangement detection kit in Example 1
[0089] 20 paraffin-embedded tissues provided by the hospital were performed using the ALK gene rearrangement detection kit (including SEQ NO.41-50 and SEQ NO.51-60, a total of 20 probes) in Example 1, and the steps were as follows. 1. Sample pretreatment process:
[0090] Preparations: Turn on the slicer (45°C-60°C); turn on the water bath (37±1°C); preheat the pepsin buffer in a 37±1°C water bath, use pepsin and preheated pepsin Prepare a working solution of pepsin with a concentration of 1.0 mg / mL in the buffer solution, and place the working solution in a water bath at 37±1°C to preheat (preheat for at least 1 hour).
[0091] Section dewaxing: Submerge the section in xylene at room temperature, dewax for 10 min, replace with fresh xylene and repeat dewaxing twice. The xylene-treated slices were submerged in absolute ethanol for 5 min, and repeated with fresh absolu...
Embodiment 3
[0123] The influence of the selection of embodiment 3 primer pair quantity on sample detection result
[0124] For the upstream and downstream of the ALK gene rearrangement breakpoint, a probe library was constructed, and the first group and the second group of amplification primer pairs of different numbers were respectively selected to design 4 groups of experiments to study different numbers of amplification products (i.e., probe Needle) to determine whether the detection effect of the corresponding probe library is consistent, in which the test group 1 selected 1 pair of amplification primers; test 2 selected 3 pairs of amplification primers; test 3 selected 5 pairs of amplification primers Right; in experiment 4, 10 pairs of amplification primers were selected for each. The specific test arrangements are shown in Table 10. The synthesis of probes, the construction of probe libraries, the labeling of probes and the experimental steps are shown in Example 1 and Example 2. ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com