Method for establishing plasmid capable of knocking out mouse CRAMP gene by use of TALEN technique
A gene and mouse technology, applied in the field of constructing knockout mouse CRAMP gene plasmids, to achieve the effect of high transfection efficiency, short cycle and accurate targeted knockout
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
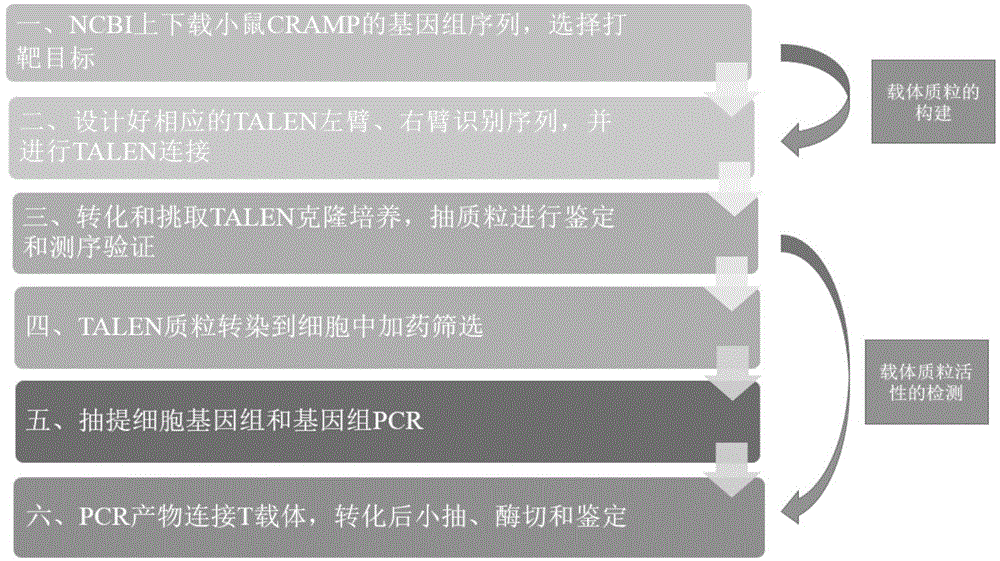
[0034] The procedure of TALEN plasmid construction and activity identification in this embodiment is shown in figure 1 .
[0035] 1. Construction of TALEN prokaryotic expression vector capable of knocking out CRAMP gene.
[0036] (1) NCBI downloaded the mouse CRAMP genome sequence (SEQ ID NO.3) and selected the target. target sequence:
[0037] atgcagttccagagggacgtcccctccctgtggctgtggcggtcactatcactgctgctgctactgggcctggggtctcccagaccccagctacagggatgctgtgctccgagctgtggatgacttcaaccagcagtccctagacaccaatctctaccgtctcctggacctggatcctgagccccaaggg.
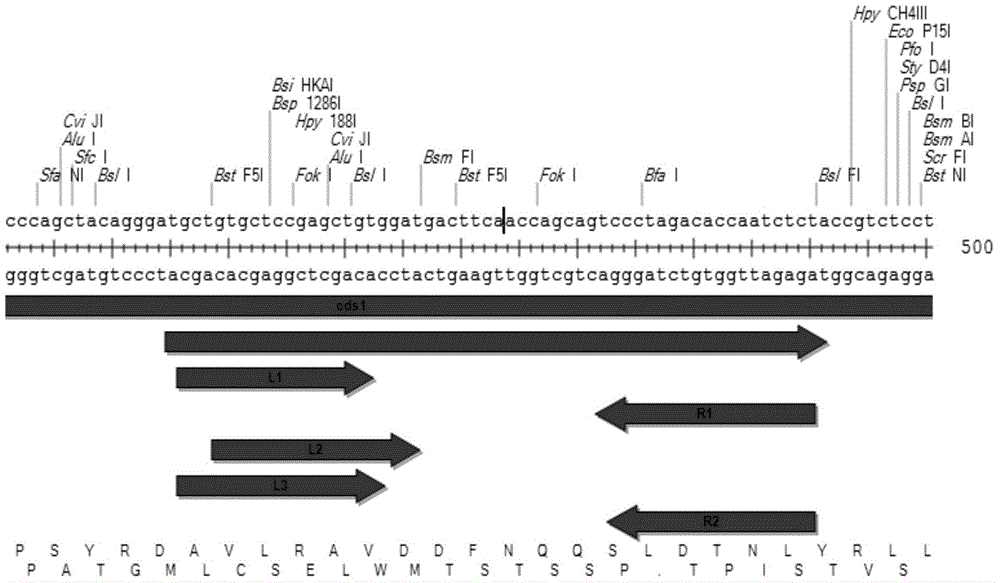
[0038] (2) According to the TALENs design principle, design in the TALENs sequence design software (https: / / tale-nt.cac.cornell.edu), find the CDS region of the CRAMP gene from the example table, and design the corresponding TALEN left arm 3 recognition sequences, 2 TALEN right arm recognition sequences. See the design results figure 2 and Table 1.
[0039]Table 1 Design of left and right arms of CRAMP gene CDS1TALENs
[0040] na...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com