Molecular ID and identification method of cordyceps sinensis
A molecular ID card, Cordyceps sinensis technology, applied in the field of identification of Chinese medicinal materials source varieties, can solve the problems of high subjectivity, poor versatility, and high requirements for observer experience, and achieve the effect of wide applicability and accurate identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
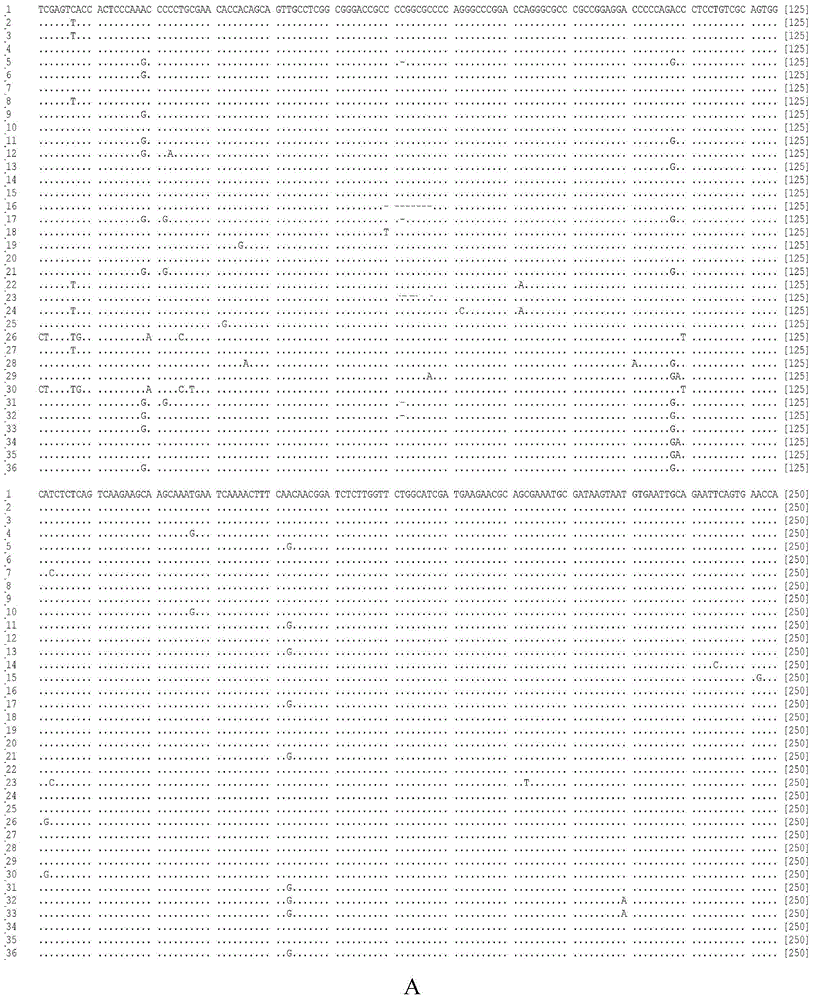
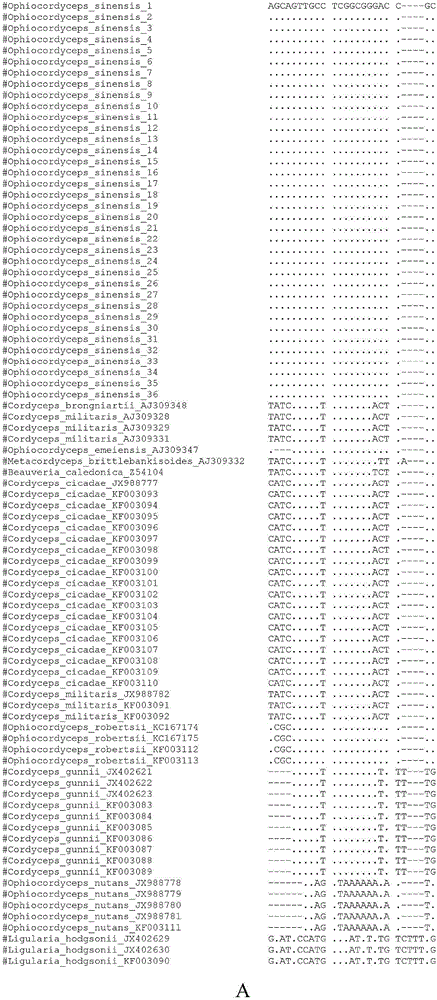
[0032] 1) 53 samples of Cordyceps sinensis O.sinensis and its obfuscated products C.gunnii, Cicadae, C.militaris, O.nutans, and O.robertsii were collected from different origins, respectively. A 20 mg sample was ground on a MM400 ball mill (Retsch, Germany), and the total DNA was extracted with a plant genomic DNA kit from Tiangen Biochemical Technology (Beijing) Co., Ltd.
[0033] 2) PCR amplification
[0034] The primer sequence is ITSF 5'-GGAAGTAAAAGTCGTAACAAGG-3'; ITSR 5'-TCCTCCGCTTATTGATATGC-3', synthesized by Sangon Bioengineering Co., Ltd. (Beijing). Primers were dissolved in sterile deionized and diluted to 2 μmol / μL. 25μL reaction system: LATaq PCR buffer II (Mg 2+ Plus) (10×) 2.5μL, dNTPs mixture 1μL (2.5mmol / L), each primer 0.6μL (10μmol / L), template DNA 1μL (- about 30ng), LA Taq DNA polymerase 1.0U, plus sterilized double-distilled Water to 25 μL for PCR amplification.
[0035] PCR reaction program: Perform the amplification reaction on the PCR instrument acco...
Embodiment 2
[0044] The identification of embodiment 2 Cordyceps sinensis mycelium
[0045] Using the same method as in Example 1, the difference is that the Cordyceps sinensis mycelium is used as a sample, DNA is extracted, and identification is carried out. As a result, the above two SNP sites can also be specifically detected. And the sequence comparison found that the ITS sequence of DNA of Cordyceps sinensis decoction pieces contained AGCAGTTGCCTCGGCGGGACCGC and ACCCCGCCGCGGCTCCCCTGCGCAGT.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com