Molecular breeding method of short catkin chestnut varieties
A short male inflorescence, molecular breeding technology, applied in the fields of botanical equipment and methods, biochemical equipment and methods, and microbial determination/inspection, can solve the problems of long selection period and low selection accuracy, and improve hybridization efficiency. , the effect of reducing the amount of hybrid labor
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Development of Molecular Markers Related to Chestnut Short Stamina Characters
[0038]The SLAF-seq (Specific-Locus Amplified Fragment Sequencing) technology developed by Beijing Biomark Biotechnology Co., Ltd. was used to conduct association analysis on the genetic segregation population of chestnut short male flowers, and obtain molecular markers closely related to chestnut short male flowers. The specific process is as follows: use the chestnut genome to identify SNPs through bwa comparison software and GATK software. And filter the SNP. The filtering method is as follows: first filter the sites with a read support degree of less than 3, and then filter the sites in the offspring mixed pool that differ from the SNP types of the corresponding parents, and finally obtain high-quality credible SNP sites, and identify them on this basis The point of difference between the two pools. Finally, association analysis was carried out using the loci of differences be...
Embodiment 4
[0064] Example 4 The method of cultivating new varieties of Chinese chestnut with insect resistance and storage resistance and short male inflorescence
[0065] The method comprises the steps of:
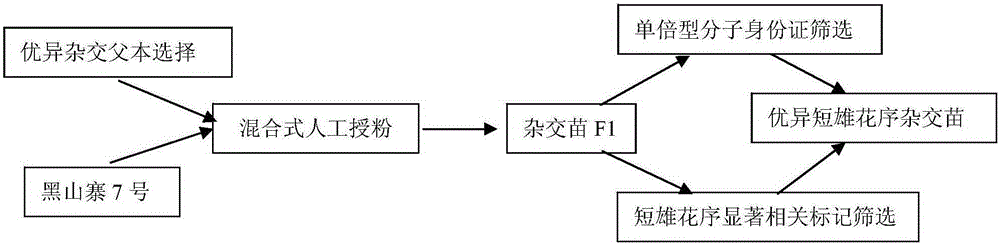
[0066] 1) Configure the hybrid combination: take the chestnut variety 'Heishanzhai No. 7' as the female parent, and select excellent germplasm resources such as 'Extra Early Maturity', 'Dayouli', 'Gushi No. 4' as the male parent, collect pollen and mix 'Heishanzhai 7' was pollinated. In the pollination process, 'Heishanzhai 7' did not need to be detasseled and bagged. The true hybrids were screened with primer I, and the F1 generation was obtained by hybridization.
[0067] 2) Constructing the gene haplotype molecular ID card of the hybrid parent: same as step 2) of Example 2.
[0068] 3) Molecular marker screening of hybrid seedlings: using the 'Gushi No. 4' haplotype molecular ID card and the molecular markers developed in Example 1 related to the chestnut short male inflorescenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com