Methane/ ammoxidation bacterial community structure analysis method based on two-step method
A technology of ammonia oxidizing bacteria and analysis methods, which is applied in the directions of biochemical equipment and methods, and the determination/inspection of microorganisms, which can solve the problems of low primer amplification efficiency, improve primer coverage, ensure PCR efficiency, and maintain PCR efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
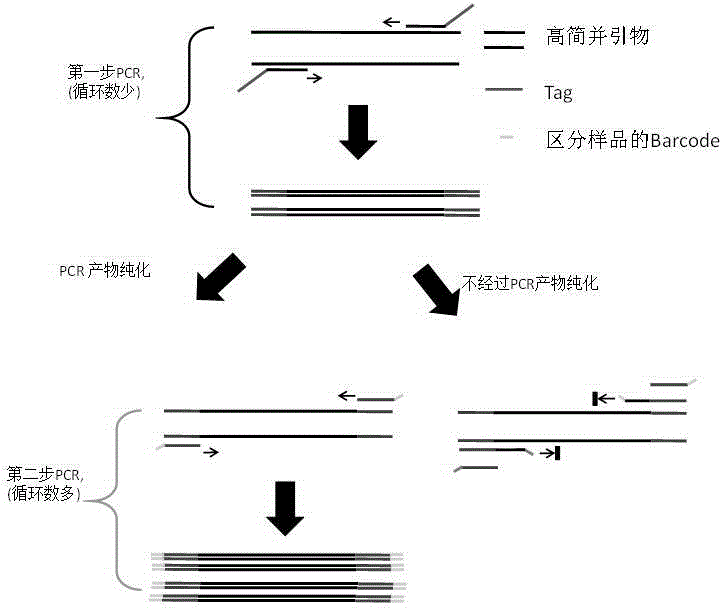
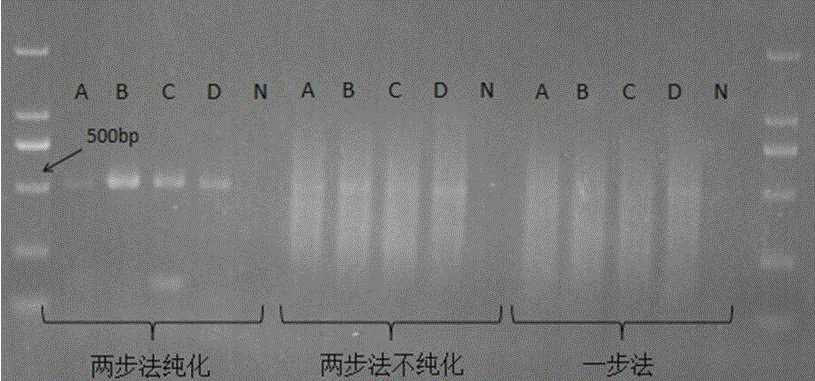
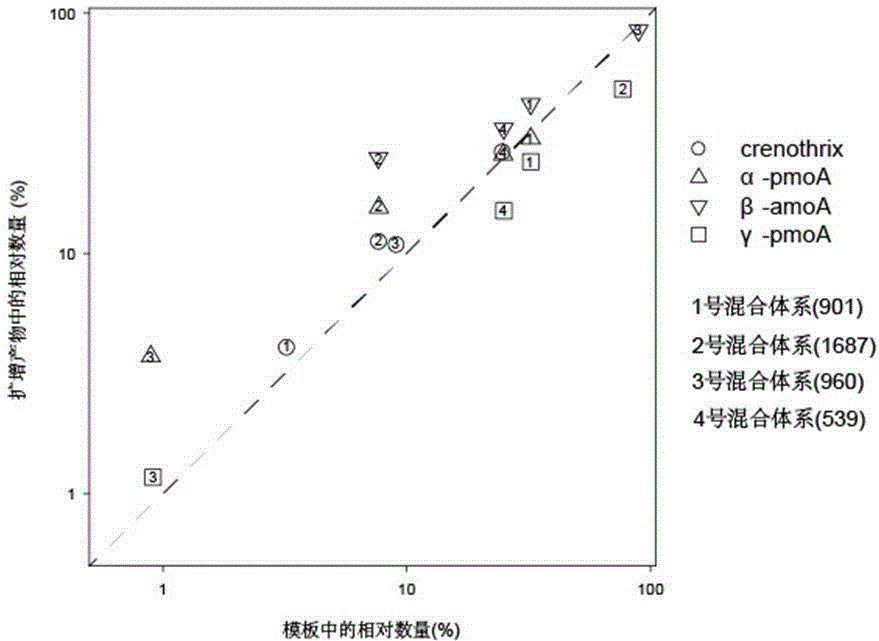
[0031] Analysis of Methane Oxidation-Associated Microbial Diversity in Multiple Environmental Samples Using Two-Step PCR.
[0032] 1. Design primers for the first step of PCR, that is, highly degenerate tagged primers for pmoA / amoA related genes (primer direction 5'→3')
[0033] forward:A189-tag:GCCGGAGCTCTGCAGATATCGGNGACTGGGACTTCTGG (SEQ ID NO.1);
[0034]reverse: mb616-tag: GCCGGAGCTCTGCAGATATCAYCWKVCKNAYRTAYTCVGG (SEQ ID NO. 2).
[0035] 2. Design primers for the second step of PCR, namely tag
[0036] tag: (5'-GCCGGAGCTCTGCAGATATC-3') (SEQ ID NO.3) Note: The forward and reverse primers in the second step of PCR are the same, and both are tag sequences.
[0037] 3. The first step PCR system and procedure
[0038] PCR kit 2×TaqPCR MasterMix (TIANGEN);
[0039] A 50 μL system was used for PCR amplification, including: 25 μL of 2×TaqPCR MasterMix, primers A189f-tag and mb616-tag (final concentration 0.2 μM), and genomic DNA from environmental samples.
[0040] The PCR ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com