A kind of emsa method and its probe and the preparation method of the probe
A probe and linker sequence technology, which is applied to the EMSA method and the probe and the preparation field of the probe, can solve the problems of the proportion of double-stranded probes affecting the development of positive results, the difficulty of guaranteeing the proportion of double-stranded probes, and false negatives. , to achieve the effect of shortening the marking time, reducing the preparation cost and overcoming the development error
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
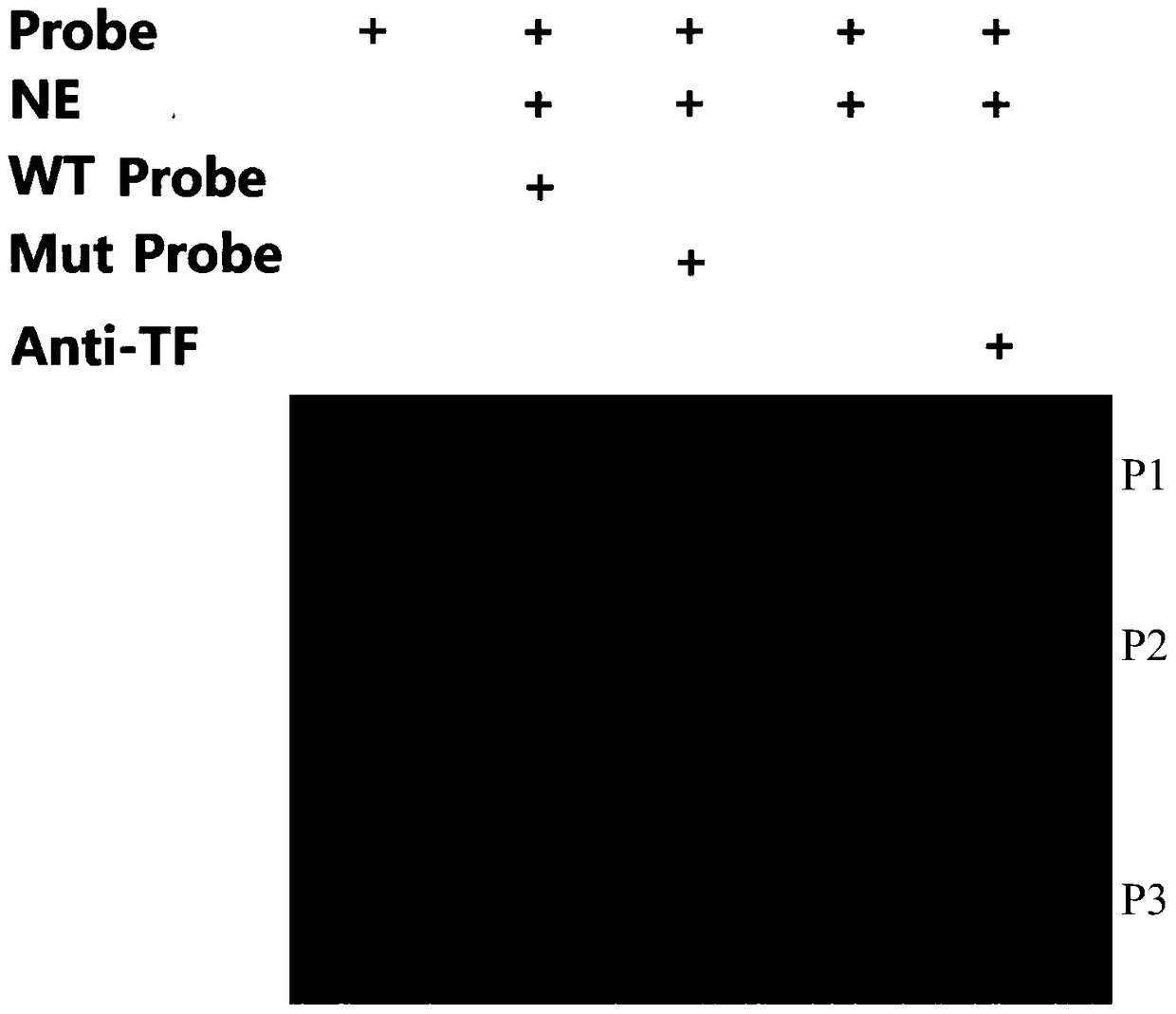
Examples
Embodiment 1
[0042] Example 1, SMAD2
[0043] S1. Preparation of template chain
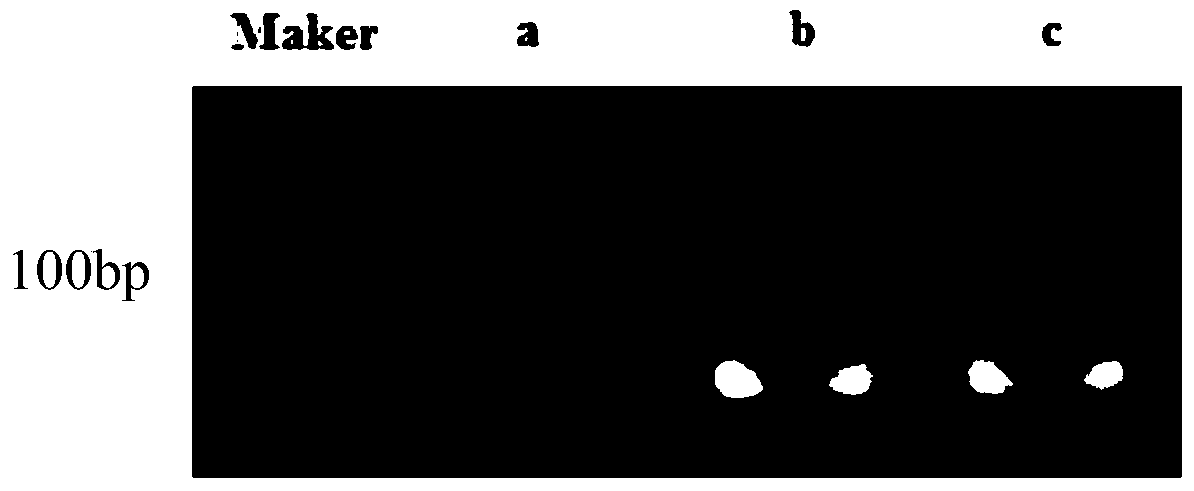
[0044] The transcription factor binding sequence was directly determined according to the SMAD2 sequence of HOCOMOCO or JASPAR database, and the obtained transcription factor binding sequence was GTGTCHGKCTR (H=A / C / T; K=G / T; R=A / G). A linker sequence (GGGTCTAGACCC, SEQ ID NO: 1) was added to the 5' and 3' ends of the transcription factor binding sequence to obtain a template strand (linker sequence + transcription factor binding sequence + linker sequence, GGGTCTAGACCC+GTGTCHGKCTR+GGGTCTAGACCC, SEQ ID NO: 4), its structure is as figure 1 As shown, the sequence was sent to Gene Synthesis Company for synthesis. Simultaneously, a mutant single chain was synthesized with the sequence GGGTCTAGACCC+TCGRTTHGKCG+GGGTCTAGACCC (SEQ ID NO: 7).
[0045] S2. Synthetic primers
[0046] Chemically synthesize biotin-labeled and LNA-modified primers. The nucleotide sequence of the primer is the same as that of the linker ...
Embodiment 2
[0079] Embodiment 2, NFKB3
[0080] According to the NFKB3 sequence of HOCOMOCO or JASPAR database, the transcription factor binding sequence was directly determined, and the obtained transcription factor binding sequence was AGTTGGAAATYCCTCCCAGGC. A linker sequence (GCATCATGATGC, SEQ ID NO: 2) was added to the 5' and 3' ends of the transcription factor binding sequence to obtain a template strand (linker sequence + transcription factor binding sequence + linker sequence, GCATCATGATGC+AGTTGGAAATYCCTCCCAGGC+GCATCATGATGC, SEQ ID NO: 5). The sequence was sent to Gene Synthesis Company for synthesis. Simultaneously, a mutant single chain was synthesized, the sequence of which was GCATCATGATGC+AGCTAGYCTGACATCACGCTG+GCATCATGATGC (SEQ ID NO: 12).
[0081] The methods of synthesizing primers, PCR amplification and synthesizing double-stranded probes and EMSA in this example are the same as those in Example 1. The sequence of the primer modified by LNA is GcATcATgATgC (lowercase let...
Embodiment 3
[0086] Example 3, C-JUN
[0087] The transcription factor binding sequence was directly determined according to the C-JUN sequence of HOCOMOCO or JASPAR database, and the obtained transcription factor binding sequence was TCCGTGTTCTGACTCTTGAGGGTCTTC. A linker sequence (GGGCTAGCCC, SEQ ID NO: 3) was added to the 5' and 3' ends of the transcription factor binding sequence to obtain a template strand (linker sequence + transcription factor binding sequence + linker sequence, GGGCTAGCCC+TCCGTGTTCTGACTCTTGAGGGTCTTC+GGGCTAGCCC, SEQ ID NO: 6). The sequence was sent to Gene Synthesis Company for synthesis. Simultaneously synthesize a mutant single chain, the specific sequence of which is GGGCTAGCCC+TCCTCGTCGTGTCAGTGGTCTTATGTC+GGGCTAGCCC (SEQ ID NO: 17).
[0088] The method of synthesizing primers and PCR amplification and synthesizing double-stranded probes in this example is the same as that in EMSA Example 1, but the sequences are different. The sequence of the primer modified by...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com