A feature analysis method for RNA editing sites
A technology of RNA editing and feature analysis, applied in the field of feature analysis of RNA editing sites, can solve the problems of energy investment, lack of analysis ideas, software and hardware support, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] 1. Sample / data source
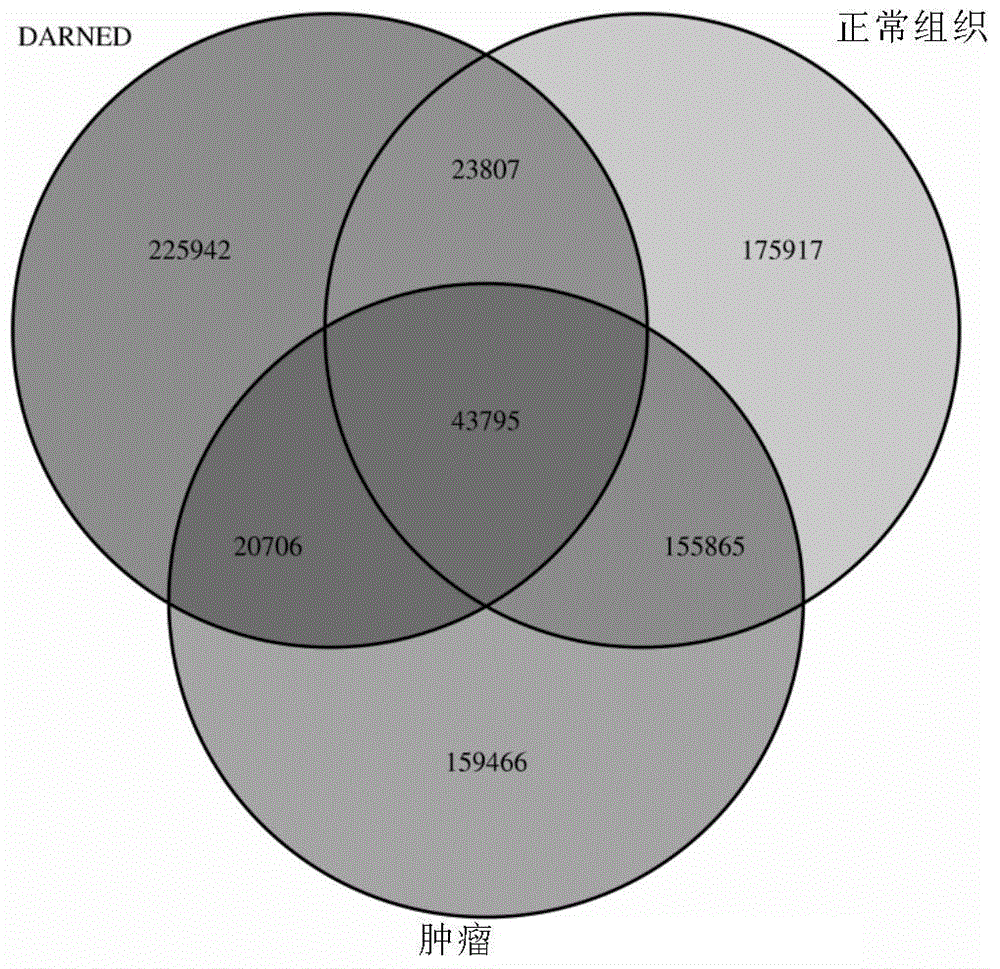
[0095] 1. In 165 prostate cancer patients, high-throughput sequencing was performed on the normal DNA, tumor DNA, normal RNA, and tumor RNA of each patient, with a read length of 90 bp, and the RNA editing site data and SNP site data of the population were analyzed and obtained to obtain RNA editing sites and corresponding annotation information in VCF format.
[0096] 1.2Darned database (URL: http: / / darned.ucc.ie / )
[0097] 2. Analysis and processing of RNA editing site data
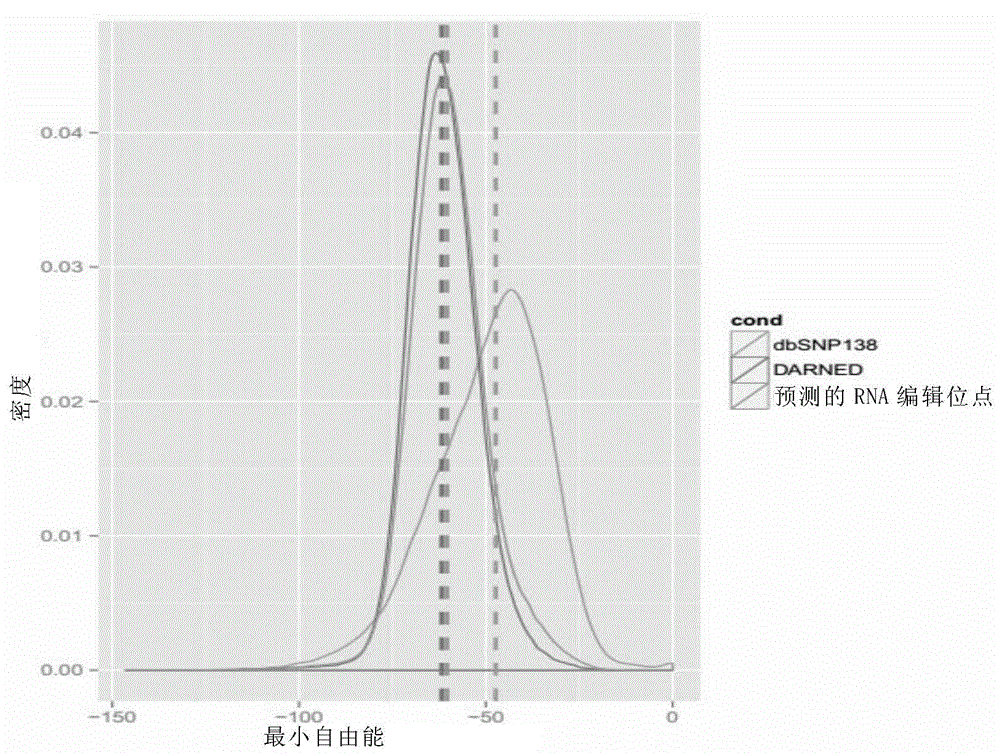
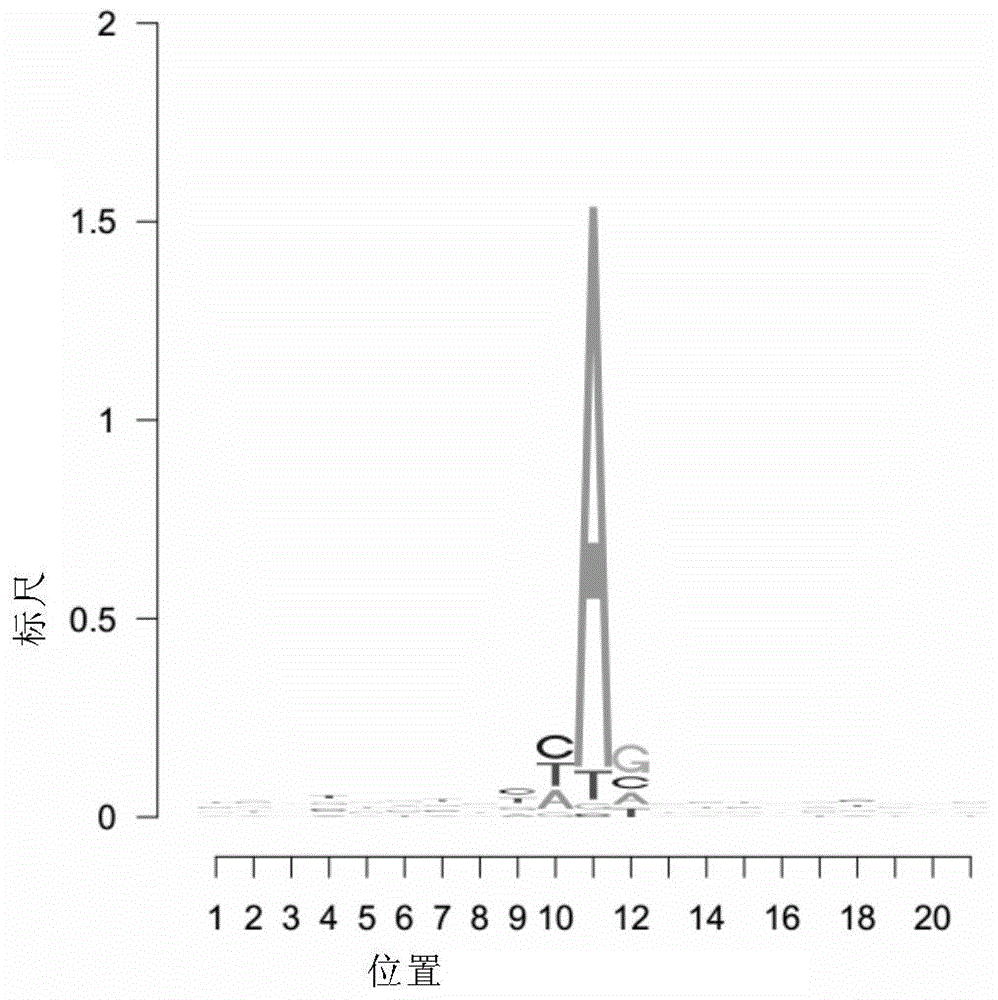
[0098] In this embodiment, RNAfold software is used to analyze the characteristics of RNA editing sites. RNAfold software is open source software, and the download address is as follows: http: / / www.tbi.univie.ac.at / RNA / index.html#download .
[0099] For the convenience of description, the generated files and descriptions in this embodiment are listed in Table 1.
[0100] Table 1 Generated files and descriptions in this embodiment
[0101]
[0102]
[0103]
...
Embodiment 2
[0119] The steps in Example 1 were repeated, with the difference that the following samples were used to replace the 65 prostate cancer patients in Example 1, so as to obtain RNA editing site data sets and perform feature analysis:
[0120] Sample: 24 patients with lung cancer.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com