Method for identifying cordate houttuynia and herb of Chinese gymnotheca
A technology of Houttuynia cordata and 100-step revival, which can be used in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1. Design of multiplex PCR primers for identifying Houttuynia cordata and Baibu Huanhun
[0060] 1. Genomic DNA extraction
[0061] Genomic DNA of Houttuynia cordata Thunb and Gymnothe cachinensis Decne were extracted by alkaline lysis method. The specific steps of the above-mentioned alkaline lysis method for extracting genomic DNA are as follows:
[0062] 1) Take 10-25mg of dry material (Houttuynia cordata or Baibu Huanhun), crush it, and put it into a 2.0ml centrifuge tube.
[0063] 2) Add 200 μL of alkaline lysis extract solution (NaOH 0.5M, PVP 1% and Triton×1001%), and vortex well to mix.
[0064] 3) Add 800 μL of neutralizing solution (Tris-HCl 0.1M, pH=8.0), shake and mix well, centrifuge at 12000 rpm for 1 min, discard the precipitate, and take the supernatant.
[0065]4) Take 500 μL of supernatant to a new 2.0ml centrifuge tube, add 500 μL of neutralizing solution (Tris-HCl 0.1M, pH=8.0), invert and mix well, centrifuge at 12000 rpm for 1 min, and t...
Embodiment 2
[0079] Embodiment 2, the method for distinguishing Houttuynia cordata and Baibu Huanhun
[0080] 1. Specific PCR to identify samples of Houttuynia cordata and Houttuyniae reincarnation
[0081] (1) Extraction of genomic DNA
[0082] Genomic DNA of Houttuynia cordata Thunb and Gymnothecachinensis Decne in Table 1 were extracted by alkaline lysis method.
[0083] Table 1. Information of experimental materials
[0084]
[0085] (2) PCR amplification
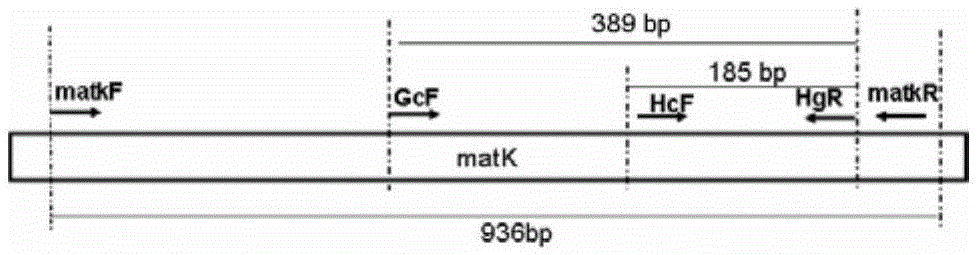
[0086] Using the genomic DNA in step (1) as a template and using primers HcF and HgR to perform PCR amplification to obtain PCR products.
[0087] PCR reaction system: a total volume of 20 μL, including 1 U of TopTaq DNA polymerase (QIAGEN, 200201), 2.0 μL of 10×buffer, 20 nmol of dNTP, 2 μL of 2 μM primers (HcF and HgR), and 20 μL of sterilized distilled water.
[0088] PCR reaction program: 94°C pre-denaturation for 3 min, 94°C denaturation for 30 s, 50°C annealing for 30 s, 72°C extension for 1 min, 72°C extension for 7 mi...
Embodiment 3
[0128] Embodiment 3, the annealing temperature sensitivity detection of multiplex PCR primer
[0129] 1. Genomic DNA extraction
[0130] Genomic DNA of Houttuynia cordata Thunb and Gymnothecachinensis Decne in Table 1 were extracted by alkaline lysis method.
[0131] 2. Multiplex PCR amplification
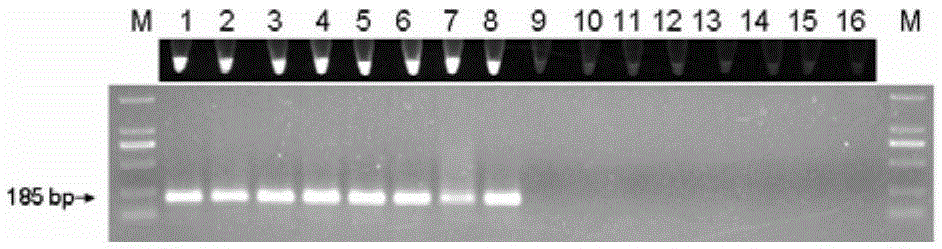
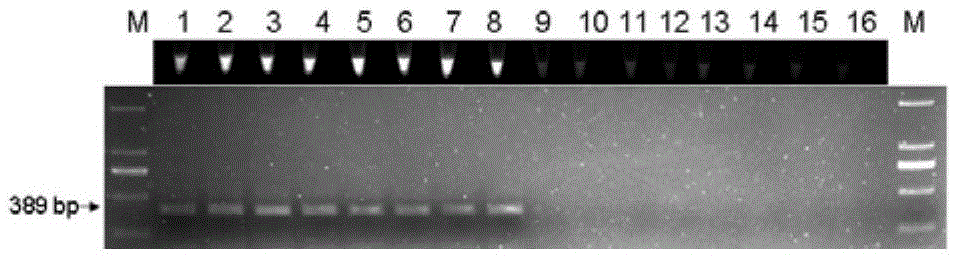
[0132] Respectively using the genomic DNA in step 1 as a template, using the method of multiplex PCR in step 2 and 3 in Example 2 to quickly identify the Houttuynia cordata and the Houttuynia reincarnation samples, and using different annealing temperatures (50°C, 50.5°C, 51.6°C , 53.2° C., 55.1° C., 56.7° C., 57.6° C., 58° C.) were subjected to PCR amplification respectively, and the remaining reaction conditions were unchanged to obtain PCR products respectively.
[0133] (3) Electrophoresis
[0134] PCR products were electrophoresed on 1.2% agarose gel. The results showed that under the condition of annealing temperature of 50-58°C (50°C, 50.5°C, 51.6°C, 53.2°C, 55.1°C, 56.7...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com