Method for enriching nucleic acids with target sequence from nucleic acid sample
A technology of target sequence and nucleic acid sequence, applied in the direction of biochemical equipment and methods, measurement/testing of microorganisms, etc., can solve problems such as no longer suitable
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Embodiment 1: the design of bait sequence
[0073] 1000 sites on the exons and introns of the human genome were randomly selected (the distribution of these sites is shown in the table) for testing the method of the present invention. Decoy sequences were designed for these 1000 random target sequences for subsequent testing.
[0074] Table 1: Chromosomal distribution of 1000 randomly selected loci
[0075] chromosome
Number
Number
chr1
92
chr12
73
chr2
67
chr13
23
chr3
53
chr14
15
chr4
43
chr15
29
chr5
45
chr16
41
chr6
124
Chr17
36
chr7
42
chr18
14
chr8
46
Chr19
31
chr9
34
chr20
21
chr10
61
chr21
9
chr11
80
Chr22
21
[0076] Decoy sequence design includes the following steps:
[0077] 1. First, the target sequence characteristic analysis includes ...
Embodiment 2
[0099] Embodiment 2: Preparation of bait sequence
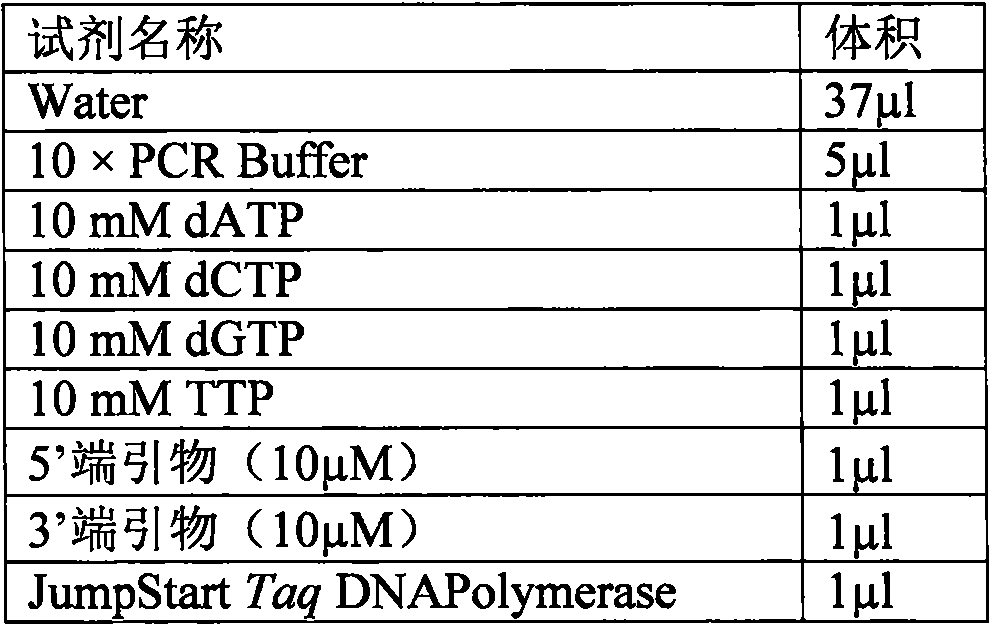
[0100] Sequence preparation was carried out according to the bait sequence designed in Example 1, and the preparation method of the bait sequence was as follows:
[0101] 1. Add specific sequences with a length of 20 bases to the 5' end and 3' end of the bait sequence respectively. The specific sequence design principles are: 1) No non-specific amplification products will be generated on the target (to be captured) genome ; 2) The GC content is between 30% and 70%, preferably between 40% and 60%; 3) The two will not form dimers, or the free energy of dimers formed is ≤47°C, preferably ≤37°C . Thus forming the sequence to be synthesized, all bait sequences are identical to a pair of specific sequences, examples are as follows:
[0102] 5' specific sequence-bait sequence (60-150bp range)-3' specific sequence is (SEQ ID NO.1):
[0103] ATATAGATGCCGTCCTAGCG-NNNNNNNNNNNNNNNNNNNNNN-TGGGCACAGGAAAGATACTT. Where "NNNNNNNNNNNN...NNNN...
Embodiment 3
[0124] Implementation 3: Target region library capture
[0125] 1. DNA library preparation for high-throughput capture sequencing:
[0126] 1) Take 1 μg of genomic DNA of the tested species, and use the ultrasonic breaker Bioruptor pico to randomly break into small fragments of 150-250bp;
[0127] 2) Use the Illumina TruSeq DNA library preparation kit for pre-capture small fragment library preparation.
[0128] 2. Use the prepared nucleic acid analog pool and the small fragment library of the target species to perform target region library hybridization capture:
[0129] 1) Preparation of blocking primers:
[0130]
[0131] Synthesize according to the above primer sequence, each synthesized at 100 OD, dilute each primer to 1000 μM, mix according to equal volume, and name it Block 1;
[0132] 2) Dilute cot-1 DNA and salmon sperm DNA to 100ng / μl, mix in equal volumes, and mark it as Block 2;
[0133] 3) Mix 6 μl Block 1 and 5 μl Block 2, labeled as Block Mix;
[0134] 4)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com