Binding molecules for bcma and cd3
A technology for binding molecules, CDR-H3, applied in the field of producing binding molecules of the present invention
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment A1
[1008] Example A1 Generation of CHO cells expressing chimeric BCMA
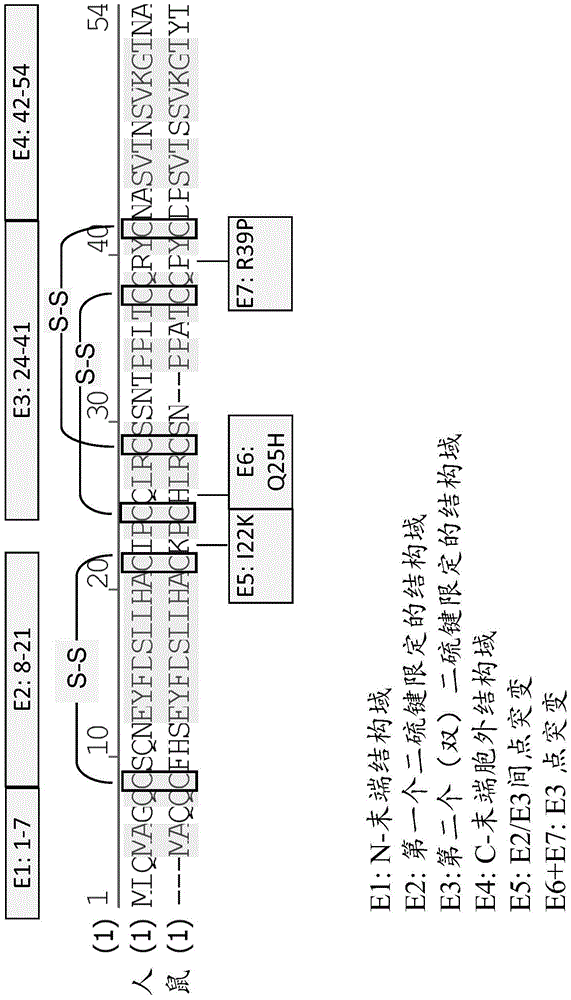
[1009] In order to construct a chimeric epitope mapping molecule, the amino acid sequence or single amino acid residue of the corresponding epitope domain of human BCMA was replaced with a murine sequence. Construct the following molecules:
[1010] ●Human BCMA ECD / E1 mouse (SEQ ID NO: 1009)
[1011] Chimeric extracellular BCMA domain: human extracellular BCMA domain, in which epitope cluster 1 (amino acid residues 1-7 of SEQ ID NO: 1002 or 1007) is divided by the corresponding murine cluster (SEQ ID NO: 1004 or 1008) Amino acid residues 1-4) replacement
[1012] →SEQ ID NO: 1002 or 1007 deletion of amino acid residues 1-3 and G6Q mutation
[1013] ●Human BCMA ECD / E2 mouse (SEQ ID NO: 1010)
[1014] Chimeric extracellular BCMA domain: human extracellular BCMA domain, in which epitope cluster 2 (amino acid residues 8-21 of SEQ ID NO: 1002 or 1007) is divided by the corresponding murine cluster (SEQ ID NO: 1004 or 1008...
Embodiment A2
[1036] 2.1 Transient expression in HEK293 cells
[1037] The clone of the expression plasmid with the nucleotide sequence of the verified sequence was used for transfection and protein expression was performed in the FreeStyle 293 expression system (Invitrogen GmbH, Karlsruhe, Germany) according to the manufacturer's instructions. The supernatant containing the expressed protein is obtained, the cells are removed by centrifugation and the supernatant is stored at -20°C.
[1038] 2.2 Stable expression in CHO cells
[1039] The clone of the expression plasmid with the nucleotide sequence of the verified sequence was transfected into DHFR-deficient CHO cells for eukaryotic expression construct. The eukaryotic protein expression in DHFR-deficient CHO cells is performed as described in Kaufman R.J. (1990) Methods Enzymol. 185, 537-566. The gene amplification of the construct was induced by increasing concentrations of methotrexate (MTX) to a final concentration of 20 nM MTX. After two ...
Embodiment A3
[1047] Example A3 Epitope clustering of mouse scFv-fragments
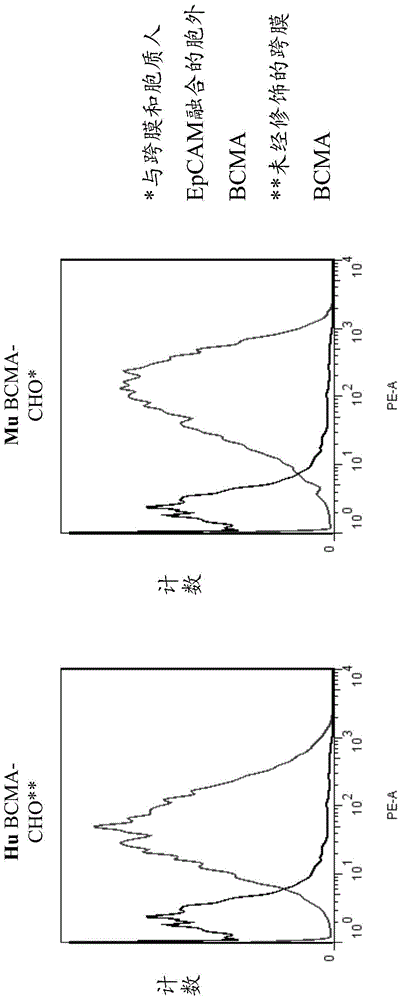
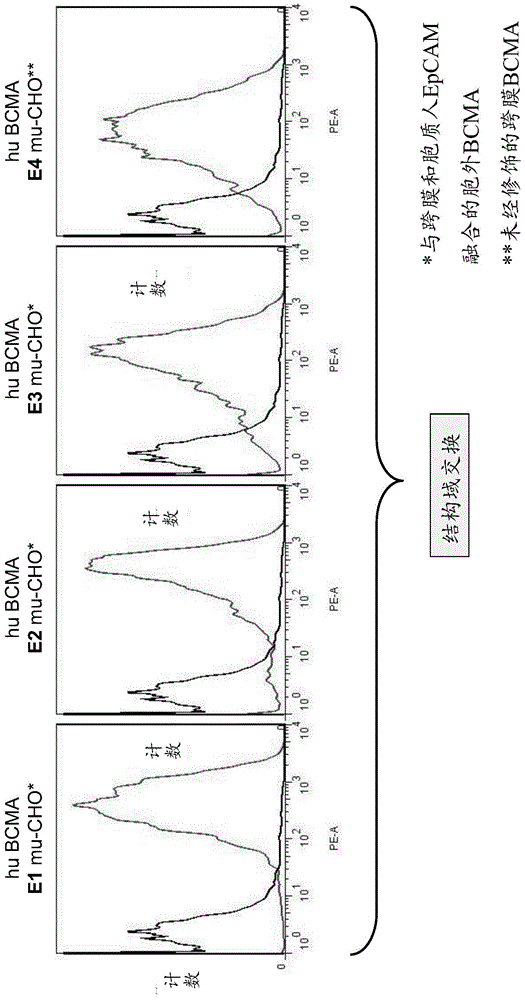
[1048] Cells transfected with human or murine BCMA, or with chimeric BCMA molecules were stained with natural, undiluted periplasmic extract containing scFv bound to human / cyno BCMA. The bound scFv was detected using 1 μg / ml anti-FLAG antibody (Sigma F1804) and R-PE-labeled anti-mouse Fcγ-specific antibody (1:100; Dianova#115-116-071). All antibodies were diluted in PBS with 2% FCS. As a negative control, cells were cultured with PBS / 2% FCS instead of periplasmic extract. The samples were measured by flow cytometry on a FACSCanto II instrument (Becton Dickinson) and analyzed with FlowJo software (version 7.6).
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Melting temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com