Distinguishing method for bacillus subtilis and bacillus amyloliquefaciens
A technology of Bacillus subtilis and Bacillus amyloliquefaciens, which is applied in the field of distinguishing similar Bacillus, can solve problems such as difficulty in distinguishing Bacillus subtilis and Bacillus amyloliquefaciens, and achieves the effects of excellent identification, accurate discrimination and simple method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0017] Specific embodiment one: the method for distinguishing Bacillus subtilis and Bacillus amyloliquefaciens in this embodiment is carried out according to the following steps:
[0018] 1. Obtain the CheA gene of the Bacillus to be identified;
[0019] 2. Comparing the homology with the CheA gene of the known type strains of Bacillus subtilis and Bacillus amyloliquefaciens, the homology lower than 80% is heterogeneous Bacillus.
specific Embodiment approach 2
[0020] Specific embodiment two: the difference between this embodiment and specific embodiment one is: Bacillus subtilis subsp.subtilis str.168 is selected as the model strain of known Bacillus subtilis in step two; Bacillus amyloliquefaciens subsp.plantarum str.FZB42 is selected as the already The type strain of Bacillus amyloliquefaciens. Other steps and parameters are the same as those in Embodiment 1.
[0021] The whole genome accession number of Bacillus subtilis subsp.subtilis str.168 is GI:728882887; the whole genome accession number of Bacillus amyloliquefaciens subsp.plantarum str.FZB42 is GI:154350369.
Embodiment 1
[0023] This example uses BLASTN 2.2.30+ to compare the homology among the 16SrRNA, GyrA, GyrB, CheA and GroEL genes of the genomes of the model strains (Bacillus subtilis subsp. subtilis str.168 and Bacillus amyloliquefaciens subsp. plantarum str. FZB42).
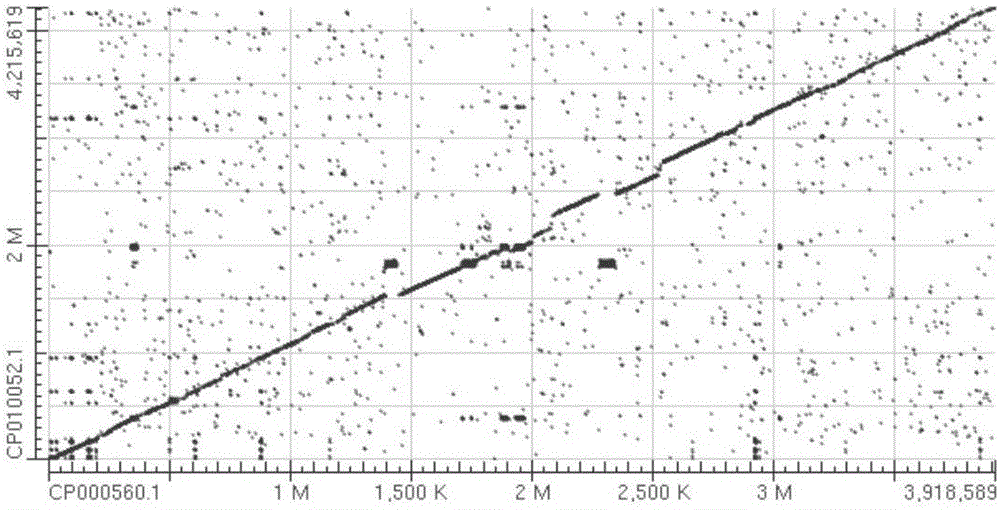
[0024] This implementation model strain comparison dot matrix analysis (Dot Matrix View) diagram as shown figure 1 shown. The results of the homology test between the genes of the model strains are shown in Table 1. The results of comparative analysis of the type strains were as follows: figure 2 shown.
[0025] Table 1
[0026] Gene
16SrRNA
GyrA
GrB
CheA
homology
99%
81%
82%
77%
93%
[0027] figure 1 and figure 2 It shows that there is a difference between Bacillus subtilis subsp.subtilis str.168 and Bacillus amyloliquefaciens subsp.plantarum str.FZB42, 80% of the nucleic acids are homologous, and the other 20% of the sequences are significantly differen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com