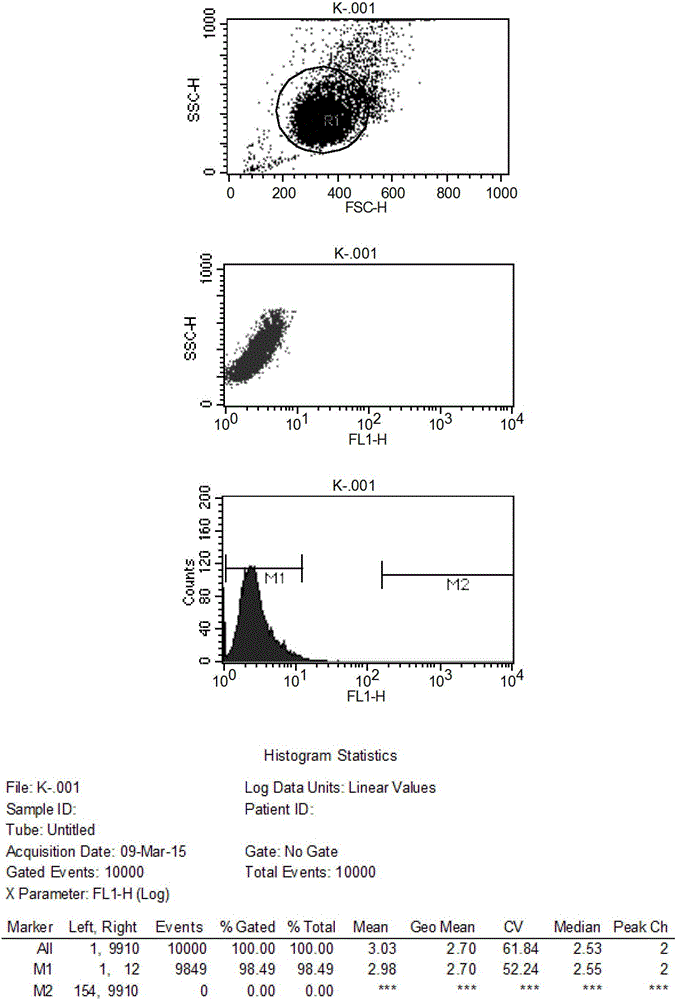
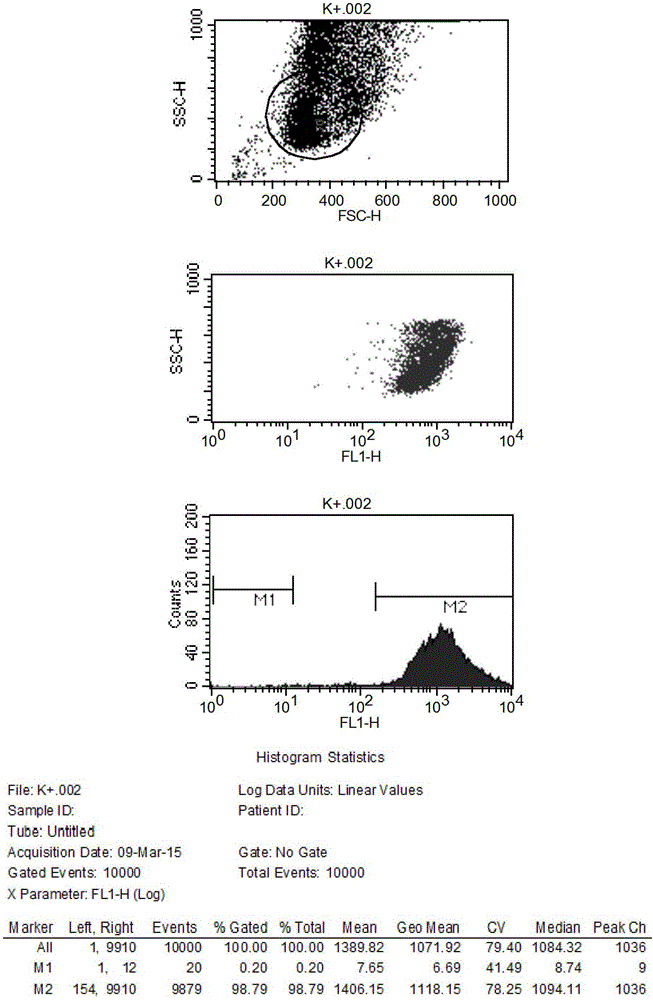
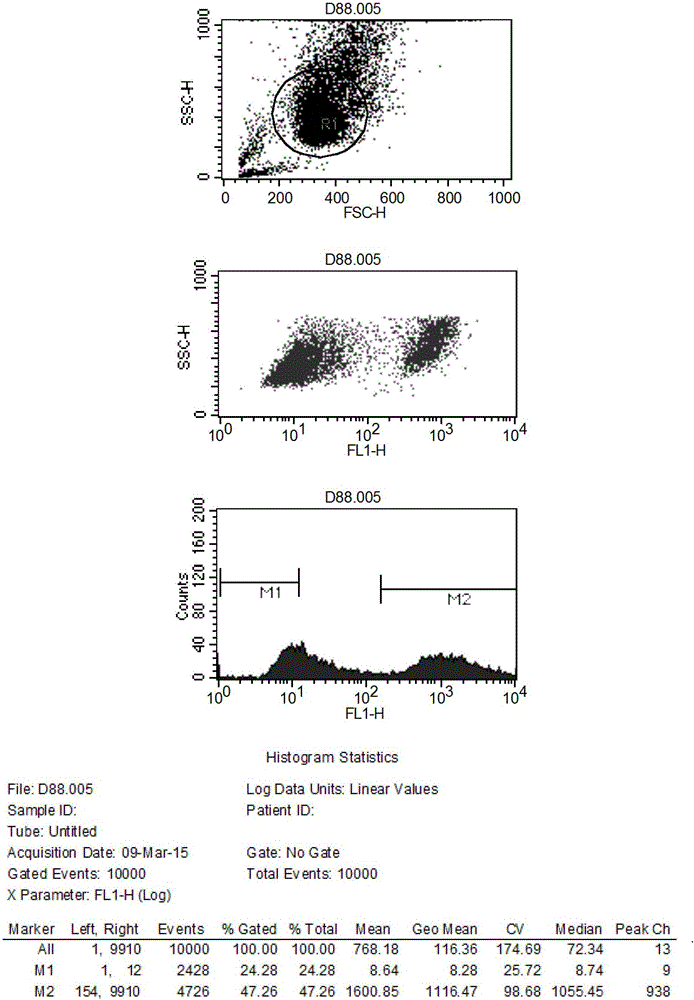
Method and application for screening ultralow fucose cell line
A fucose, host cell technology, applied in chemical instruments and methods, biochemical equipment and methods, medical preparations containing active ingredients, etc., can solve the problems of inability to stably produce antibodies and unsuitable for production of cell lines, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1: Design and vector construction of TALEN targeting FUT8 gene sequence
[0077] 1. The target information of Fut8 gene is as follows:
[0078] Fut8CDS: The yellow background marks the approximate position of TALEN cutting DNA and forming a frameshift mutation
[0079] ATGCGGGCATGGACTGGTTCCTGGCGTTGGATTATGCTCATTCTTTTTGCCTGGGGGACCTTATTGTTTTATATAGGTGGTCATTTGGTTCGAGATAATGACCACCCTGACCATTCTAGCAGAGAACTCTCCAAGATTCTTGCAAAGCTGGAGCGCTTAAAACAACAAAATGAAGACTTGAGGAGAATGGCTGAGTCTCTCCGAATACCAGAAGGCCCTATTGATCAGGGGACAGCTACAGGAAGAGTCCGTGTTTTAGAAGAACAGCTTGTTAAGGCCAAAGAACAGATTGAAAATTACAAGAAACAAGCTAGGAATGATCTGGGAAAGGATCATGAAATCTTAAGGAGGAGGATTGAAAATGGAGCTAAAGAGCTCTGGTTTTTTCTACAAAGTGAATTGAAGAAATTAAAGAAATTAGAAGGAAACGAACTCCAAAGACATGCAGATGAAATTCTTTTGGATTTAGGACATCATGAAAGGTCTATCATGACAGATCTATACTACCTCAGTCAAACAGATGGAGCAGGTGAGTGGCGGGAAAAAGAAGCCAAAGATCTGACAGAGCTGGTCCAGCGGAGAATAACATATCTGCAGAATCCCAAGGACTGCAGCAAAGCCAGAAAGCTGGTATGTAATATCAACAAAGGCTGTGGCTATGGATGTCAACTCCATCATGTGGTTTACTGCTTCATGATTGCTTATGG...
Embodiment 2
[0106] Example 2: Design and vector construction of CRISPR / CAS9 targeting FUT8 gene sequence
[0107] 1. Construction of Cas9 / gRNA, design and synthesis of targets:
[0108] Fut8 sequence information:
[0109]
[0110]
[0111]
[0112] Green background: Shearing the DNA, resulting in the approximate location of the frameshift mutation
[0113] Yellow background: Cas9 / gRNA cuts DNA, forming the approximate position of frameshift mutation
[0114]
[0115]
[0116] According to the analysis of the protein conservative region, about 180aa (located in exon 4) is the conserved region of the Fut8 super family, so exon 4 and exon 5 were selected as the DNA region for designing the gRNA target.
[0117] CHO Fut8 genome sequence: exon 4
[0118]
[0119]
[0120] Exon 5:
[0121]
[0122]
[0123] Design gRNA target for gene knockout: PAM sequence in gray background
[0124] gRNA target site position 1: CCAGCTCTGTCAGATCTT
[0125] Reverse complement:...
Embodiment 3
[0130] Example 3: Knockout of FUT8 gene in CHOK1 cells by TALEN method and sorting of monoclonal
[0131] 1. Cell culture and transfection
[0132] The following steps are applicable to mammalian cells cultured in 6-well plates. For other culture materials, please refer to the scale adjustment for transfection. All numbers and volumes are calculated by well. The ratio of DNA (ug) to Lipofectamin2000 (ul) used in most cell lines is 1:2 to 1:3, transfecting high-density cells can obtain high transfection efficiency, high expression level and low cytotoxicity. Optimal transfection is required (see DNA transfection optimization table).
[0133] 1. Suspension cells: Before preparing the transfection reagent, 2-4×10 per well 6 The cells were inoculated in 1.5 ml medium without antibiotics.
[0134] 2. Preparation of transfection reagent, the amount of cells per well is as follows:
[0135] A. Dilute plasmid DNA (2ug each of TALEN-2L and TALEN-2R plasmids) with 250ul CD OptiCHO l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com