Method and system for identifying tumor loads in samples
A sample-in-sample technology, applied in the field of identifying tumor burden in samples, can solve the problems of unstable probe hybridization efficiency, low throughput, and complicated operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
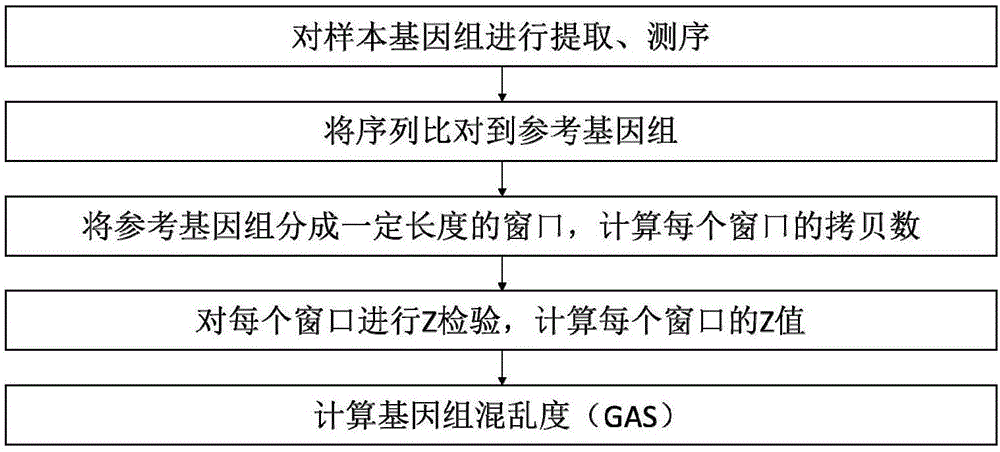
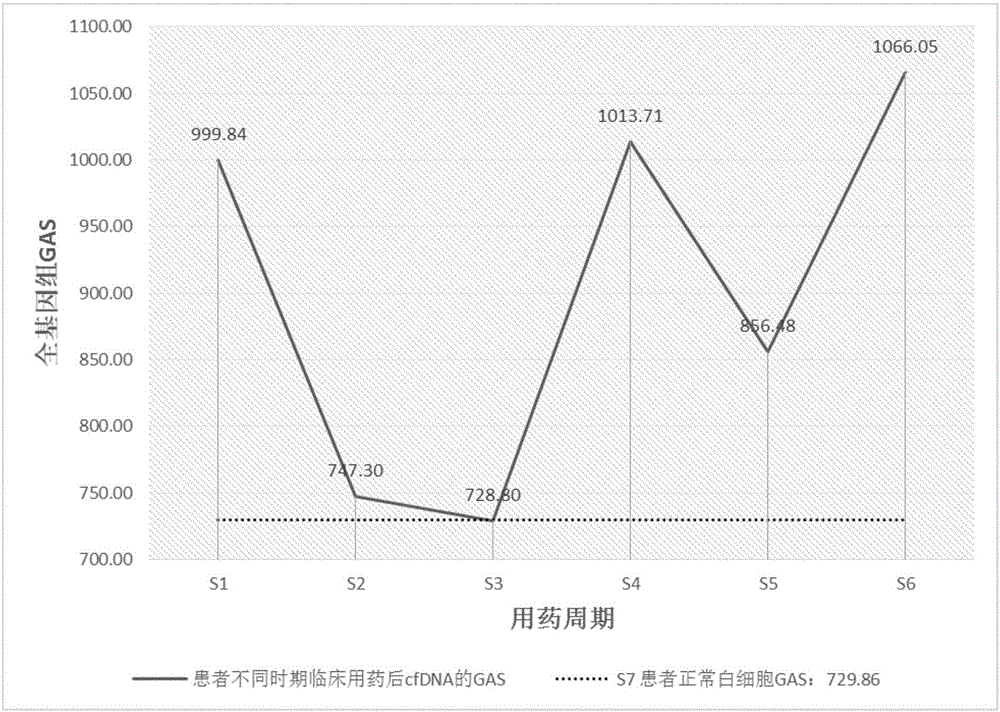
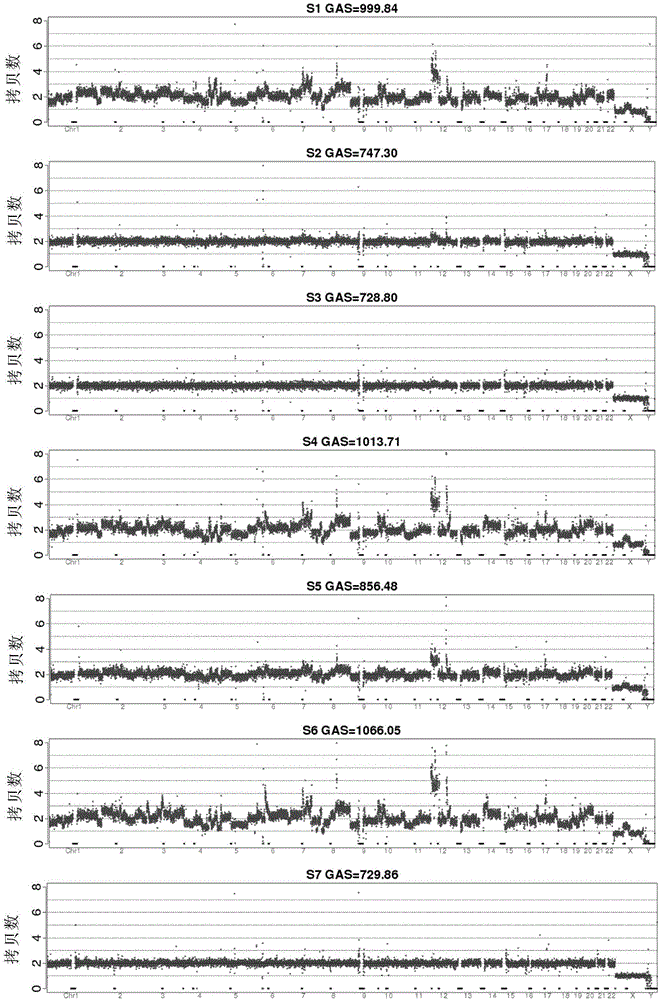
[0146] The present invention has been applied to 15 examples and has achieved good results. In order to make the usage and effects of the present invention easier to understand and grasp, an example will be given below for further elaboration. A brief flowchart of the implementation is as follows figure 1 As shown, the detailed implementation process is as follows:
[0147] 1. Nucleic acid extraction and sequencing of the sample genome
[0148] In this embodiment, the source of the detection sample is the blood of a patient with gastric cancer, and the free DNA (cfDNA) and white blood cells in the blood are extracted. Nucleic acid was extracted using the CW2603 nucleic acid extraction kit of Kangwei Century Biotechnology Co., Ltd., and the extraction method was operated according to the product manual provided by Kangwei Century Biotechnology Co., Ltd.
[0149]The CW2185 library construction kit of Kangwei Century Biotechnology Co., Ltd. was used for library construction an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com