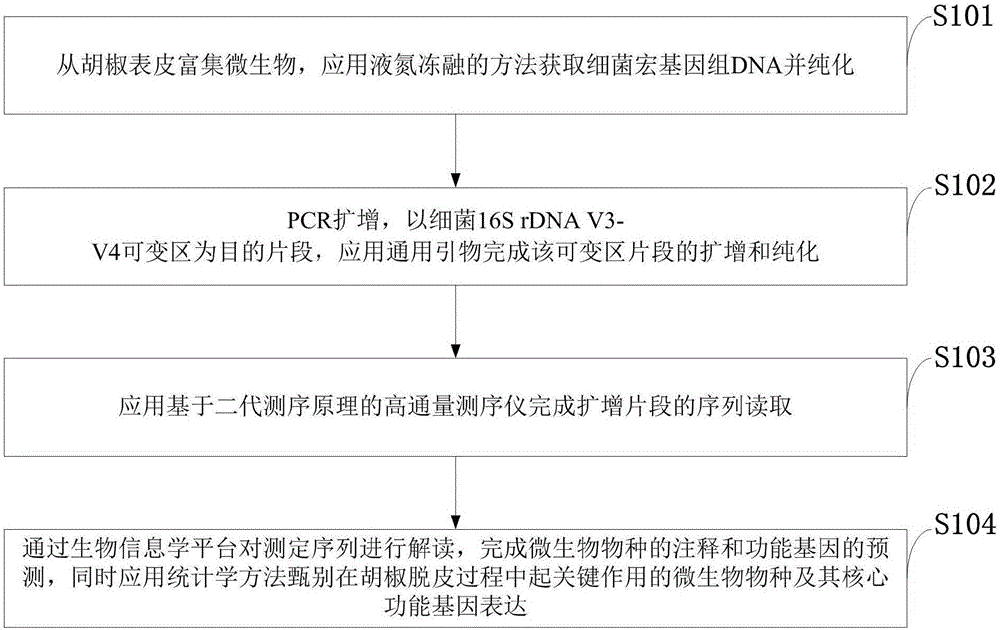
Key microbial functional genome detection method in pepper peeling process
A detection method and microbial technology, applied in the field of genetic engineering, can solve the problems of low quality pepper, peculiar smell, gray color, etc., and achieve the effects of high detection resolution and improved peeling time efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1: Detection of key microorganisms and their functional genes in the pepper peeling process in Qionghai, Hainan Province
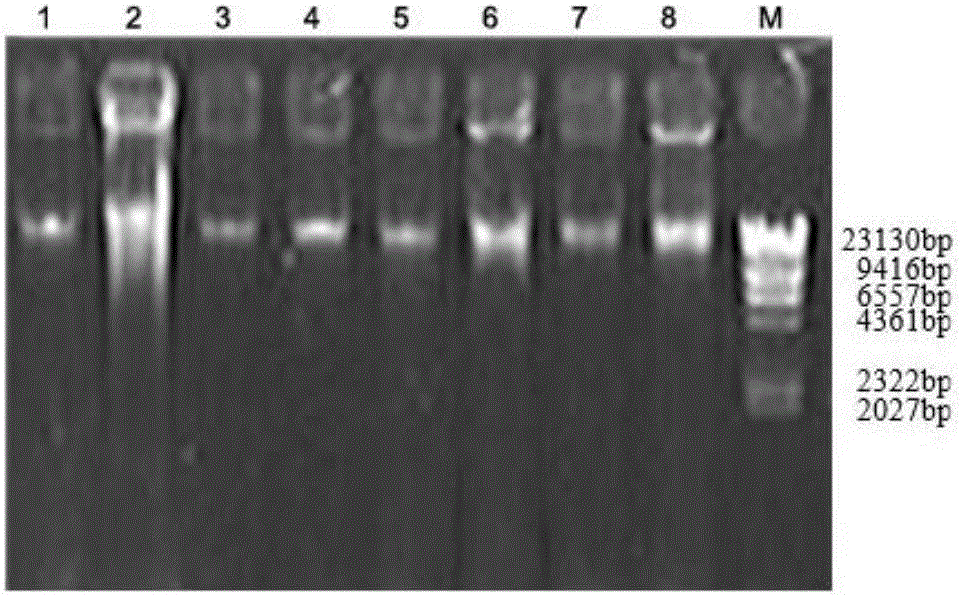
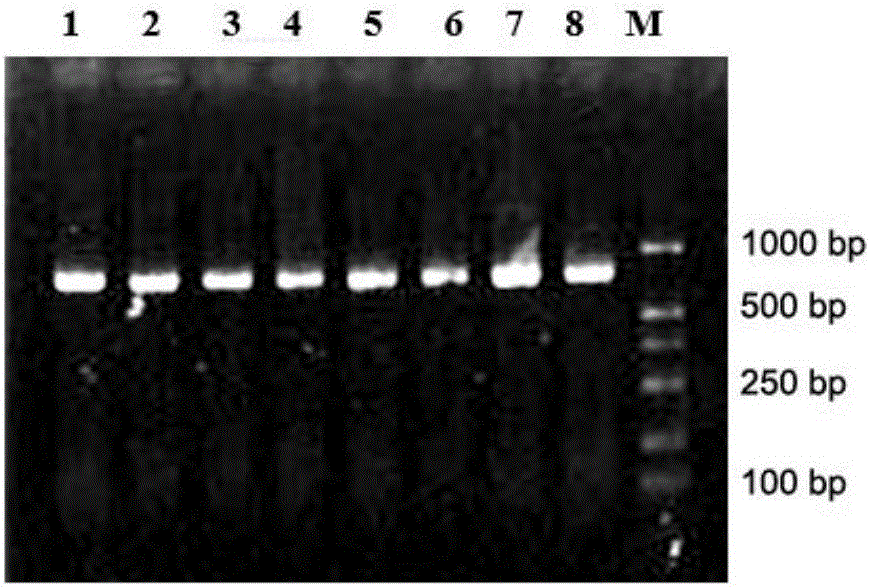
[0043] 1. Extraction of microbial metagenomic DNA from pepper epidermis:
[0044] Taking 8 pepper samples collected from the peeling stage in Qionghai, Hainan Province as the research object, before the extraction, a relatively strong liquid nitrogen freeze-thaw and ceramic bead beating and crushing method were added to release the cell contents, and then the CTAB method was used for extraction. The specific method As follows: Take 10.0g sample of pepper sample and wash it, put the eluted bacteria in a 1.5mL centrifuge tube, immediately place it in liquid nitrogen and freeze it completely, take it out and put it in a 60°C water bath to melt (about 5min), freeze and thaw repeatedly 4 times, incubate at 37°C for 4h in a constant temperature shaker, centrifuge at 12000g for 10min at room temperature, collect the supernatant and transfer to anot...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com