Nano di-cyclic aptamer probe and application thereof
An aptamer and cyclic technology, applied in the field of nano-bicyclic aptamer probes, to achieve the effects of inhibiting activity, improving specificity, and improving specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1: Design of linear aptamers
[0054] Using the tumor biomarker protein PTK7 highly expressed on the cell membrane surface of the target blood cancer cell RccF-CEM as a template, based on Cell-SELEX technology, design a linear ligand Ap that can bind to PTK7 protein with high specificity;
[0055] According to the principle of complementary base pairing, design the template T-Ap that can make the linear Ap loop closed, the linear C1 that can be complementary paired with the closed Ap partial sequence to form a small circle with a double ring structure, and the template T-C1 required for C1 loop closure, wherein , the partial sequences of the 3' and 5' ports of T-Ap and Ap are complementary, the partial sequences of the 3' and 5' ports of C1 and T-C1 are complementary, and the partial sequences of C1 and part of Ap are complementary , and, Ap contains two groups of Biotin and FAM.
[0056] The primer sequences used are as follows:
[0057] Among them, the linea...
Embodiment 2
[0065] Example 2: Transforming a linear aptamer into an aptamer probe with a functional region and a double loop region
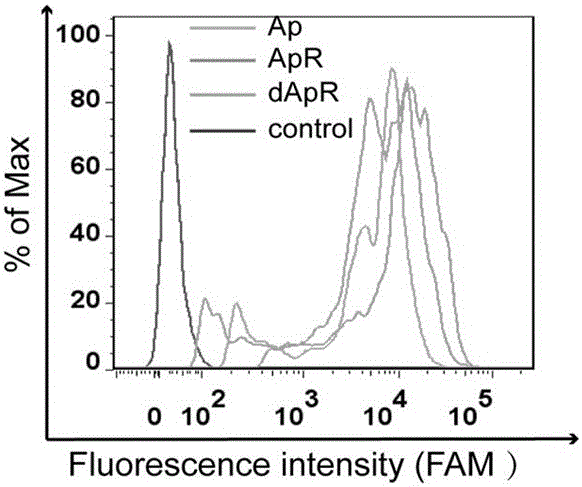
[0066] Ap diluted to 10 μM was subjected to a monocyclic annulation reaction. The reaction steps are: (1) Phosphorylation reaction was carried out by shaking reaction in a constant temperature metal bath at 37°C for 30 minutes; (2) Ligase ligation reaction was carried out by shaking reaction at 55°C for 2 hours; (3) Template removal; (4) Product purification; Finally, a single, closed circular ApR was obtained, and C1 was obtained by the same method, and C1 was smaller than ApR. Then, ApR and C1 were complementary hybridized to obtain an aptamer probe with a double-loop structure and a functional region.
[0067] Specifically include the following steps:
[0068] (1) Add linear aptamer Ap or linear oligonucleotide C1 (10nM) to the buffer system containing 1 / 3 volume of ATP and 1 / 2 volume of Polynucleotide Kinase (T4 PNK) enzyme, use dd h 2 Dilute O ten ...
Embodiment 3
[0072] Example 3: Denaturing gel electrophoresis verifies the successful construction of aptamer probes with functional regions and double loop regions
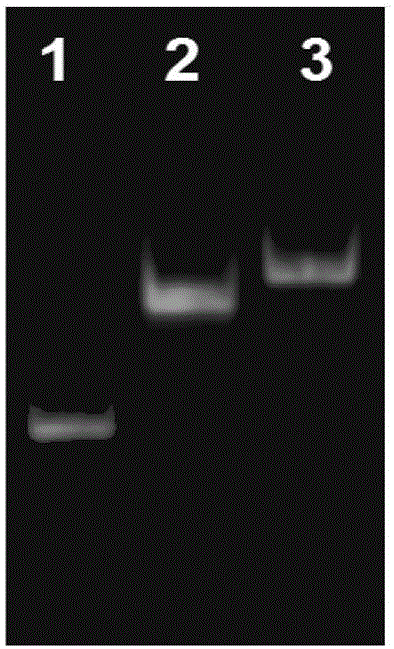
[0073] The first part is to use denaturing gel electrophoresis to verify the successful synthesis of the single-ring closed product. First, prepare a 10% denaturing bulk gel (Urea-PAGE), and combine the synthesized single-ring aptamer ApR and the double-ring structure construction. The inner circle C1, and the corresponding linear single-stranded DNA, were subjected to denaturing gel electrophoresis under the same conditions, and the single-circular aptamer was verified according to the conclusion that the swimming speed of circular DNA is slower than that of linear DNA Whether the build job for is successful. like figure 1 As shown, the band of circular DNA is significantly higher than that of linear DNA, indicating that the swimming speed is slower, which proves the successful construction of single circular DNA.
[0074]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com