Primer composition and kit for detecting autophagy key gene expression level in Drosophila melanogaster, and use method of kit
A technology of primer composition and autophagy, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/testing, etc., can solve the problems of inaccurate quantification, low sensitivity, high technical cost, etc., and achieve production saving Cost and detection cost, good sensitivity and specificity, avoid the effect of low specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The invention is a kit for detecting the expression level of key genes of autophagy in Drosophila melanogaster, and the kit includes the following reagents:
[0042] 1) DEPC water
[0043] 2) 5×RT buffer
[0044] 3) RT primer mix
[0045] 4) RT enzyme (full name reverse transcriptase)
[0046] 5) Solution Z
[0047] 6) 10×PCR buffer
[0048] 7) PCR primers
[0049] 8) 25mM magnesium chloride solution
[0050] 9) DNA polymerase
[0051] 10) Positive control
[0052] The above reverse transcription primers include RT amplification primers of 10 key genes of autophagy and RNA internal reference in Table 1 and Table 2, and the PCR primers include 10 key genes of autophagy and RNA in Table 1 and Table 2. The PCR amplification primers and gene sequences of the internal reference are shown in Table 2 below.
[0053] The above Z solution includes deoxynucleotide triphosphates (dNTPs) and universal primers. The sequence of the forward amplification primer of the univers...
Embodiment 2
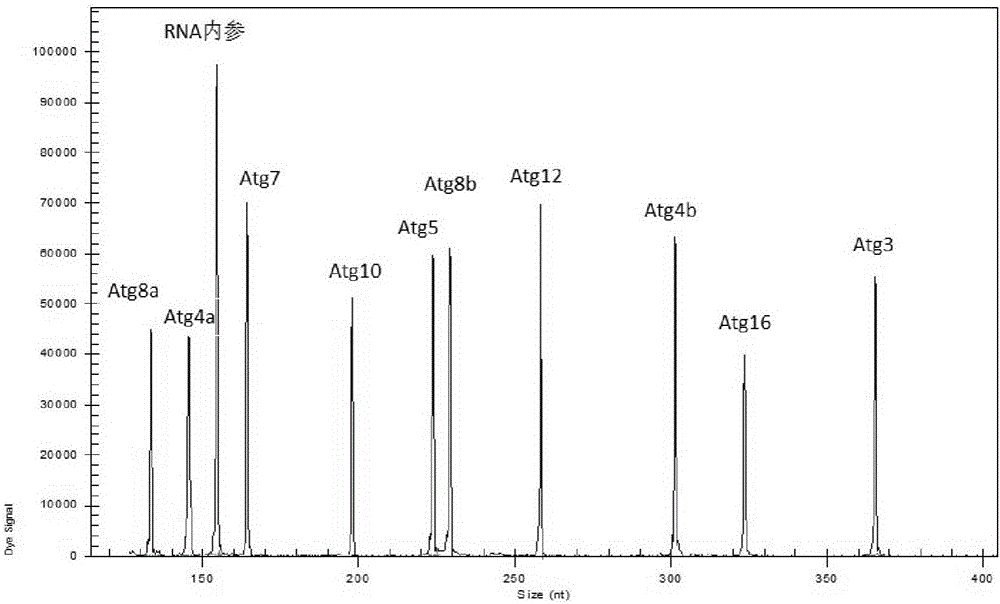
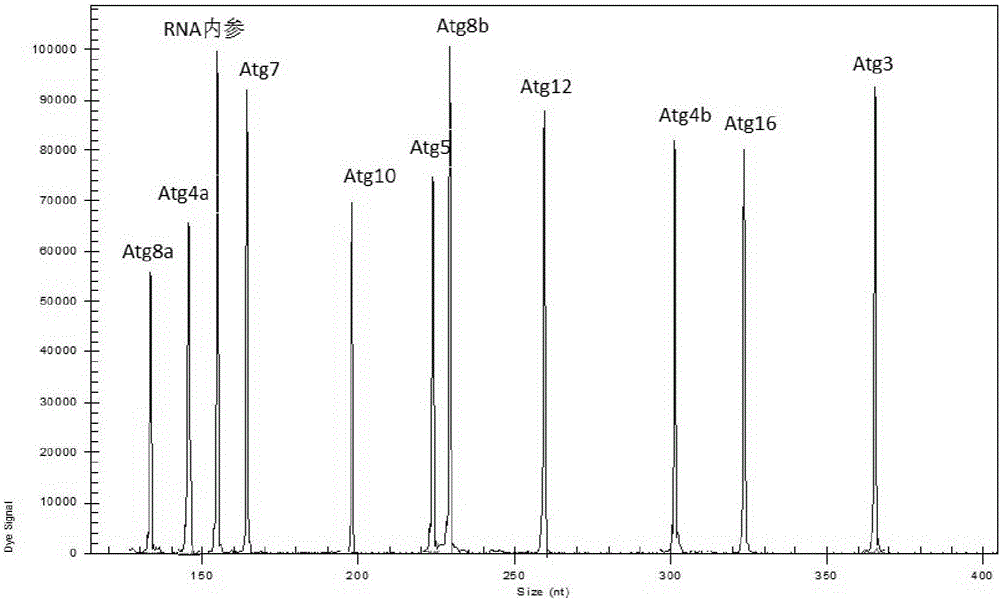
[0068] The present invention is a kit for detecting the expression level of autophagy key genes in Drosophila melanogaster, the detected genes include Drosophila melanogaster genes Atg3, Atg4a, Atg4b, Atg5, Atg7, Atg8a, Atg8b, Atg10, Atg12, Atg16 ( See Table 1). Drosophila melanogaster samples were collected, nucleic acid was extracted, reverse transcription and PCR reactions were performed using the sample nucleic acid as a template, and finally the sample was separated by capillary electrophoresis. The specific steps were as follows:
[0069] 1. Production of a kit for detecting the expression level of key genes of autophagy in Drosophila melanogaster based on the GeXP multiple gene expression genetic analysis system. The components included in the kit are the same as those in Example 1 above;
[0070] 2. Collect samples and extract nucleic acids
[0071] Collect samples from the experimental group and control group of Drosophila melanogaster, isolate and extract nucleic ac...
Embodiment 3
[0098] Embodiment 3: detection kit sensitivity, specificity analysis
[0099] Sensitivity analysis: After diluting the positive control according to a certain copy number ratio, PCR amplification and capillary electrophoresis detection until no signal is detected, the copy number is the lowest detection line, which is the sensitivity of the kit. Sensitivity up to 45 copies.
[0100] Specificity analysis: Single-plex PCR amplification is detected as a single peak of the target fragment size by capillary electrophoresis.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com