Duplex-seq-based ultralow-frequency mutation site detection analysis method
A technology of mutation sites and analysis methods, applied in the field of second-generation high-throughput sequencing, can solve problems such as ineffective presentation of data information, insufficient system of annotation process and related statistics, and no systematic analysis, so as to increase diversity , increase the quantity, improve the effect of analysis efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
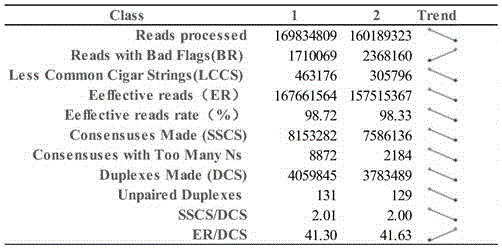
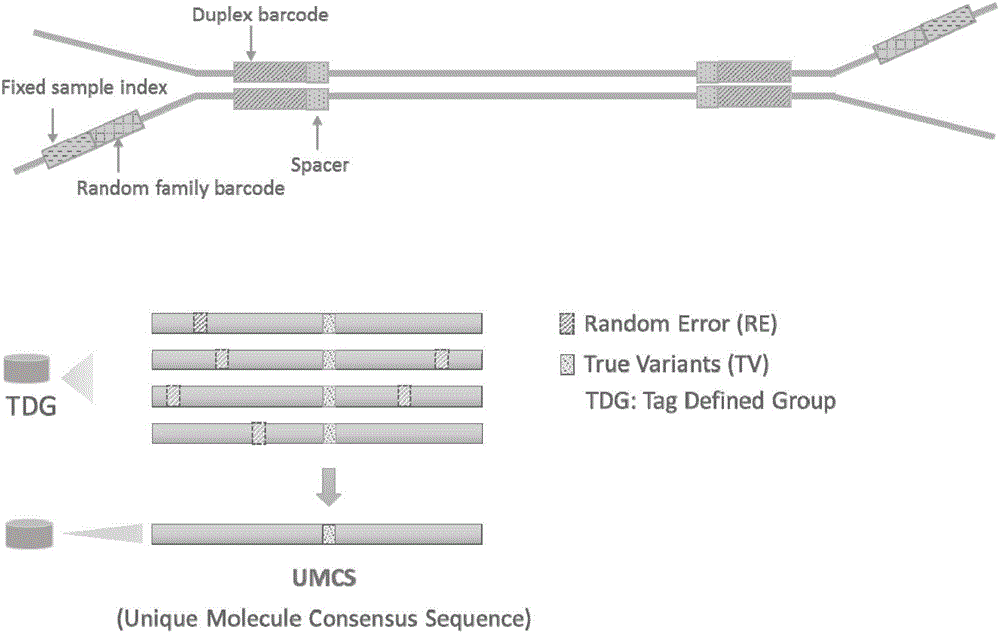
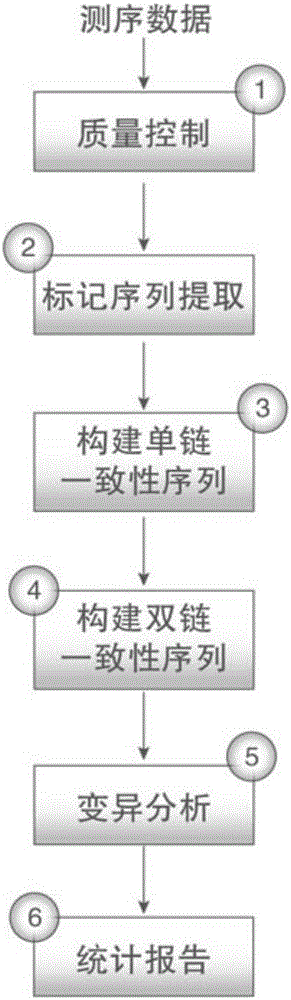
[0074] In order to realize the object of the present invention, as figure 1 and figure 2 As shown, the present invention is based on the duplex-seq ultra-low frequency mutation site detection and analysis method, comprising the following steps:
[0075] 1) Evaluate the quality of the original sequencing data, reduce data noise, and provide effective data for subsequent analysis;
[0076] 2) Extract the random barcode to the title line of each sequence in the sequence file, so as to facilitate the subsequent quick retrieval of the barcode and create a consistent sequence;
[0077] 3) Create consensus sequences based on family barcode and duplex barcode, excluding mutations introduced during library construction or PCR;
[0078] 4) Construct a double-strand consensus sequence according to the duplex-tag, and further exclude asymmetric mutation sites in the sequence;
[0079] 5) Perform local quality correction on the compared data, and detect low-frequency variant sites; ann...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com