Molecular marker DNdCAPS8.03-1 chained with corn head smut-resistant secondary and primary loci and application thereof
A head smut resistance and molecular marker technology, applied in the field of genetic engineering, can solve the problems of lack of main effect and single disease resistance gene, etc., and achieve the effect of accelerating the process of screening disease-resistant varieties.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 The establishment of the SNP site linked to the sub-major locus of resistance to head smut in maize and the molecular marker DNdCAPS8.03-1 based on this site
[0035] 1. Bioinformatics analysis of the candidate gene GRMZM2G047152 located in bin8.03 sub-main disease resistance region of maize
[0036] 1 Materials and methods
[0037] The research object of this experiment is the candidate gene GRMZM2G047152 located in bin8.03 region, which is a disease resistance gene containing NBS structure.
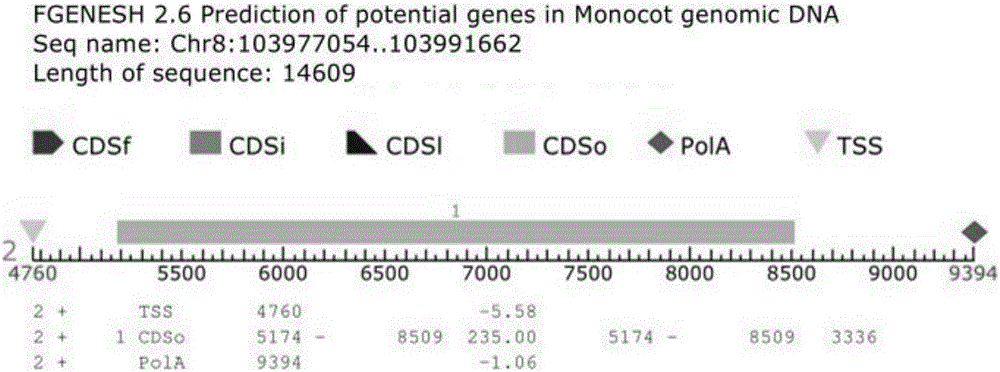
[0038] The gene GRMZM2G047152 was retrieved on the MaizeGDB and Phytozome11 websites, and the gene structure analysis of the nucleotide sequence was carried out by using the online prediction gene software Fgenesh.
[0039] The amino acid sequence of the candidate gene GRMZM2G047152 was predicted and its conserved domains were analyzed using ORF Finder and CD-search in NCBI.
[0040] Log in to the NCBI website and use the BLAST tool on the website to perform homology ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com