Target gene, primer and probes for detecting and identifying cryptococcus and kit
A cryptococcus and target gene technology, applied in the field of medical mycology, can solve problems such as the difficulty in identifying Cryptococcus neoformans and Cryptococcus gattii, the inability to distinguish between two pathogenic cryptococci, and the laboratory can only report cryptococci, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Target genes, primers, probes and kits for the detection and identification of Cryptococcus neoformans and Cryptococcus gattii
[0038] Step 1: Selection of target gene location
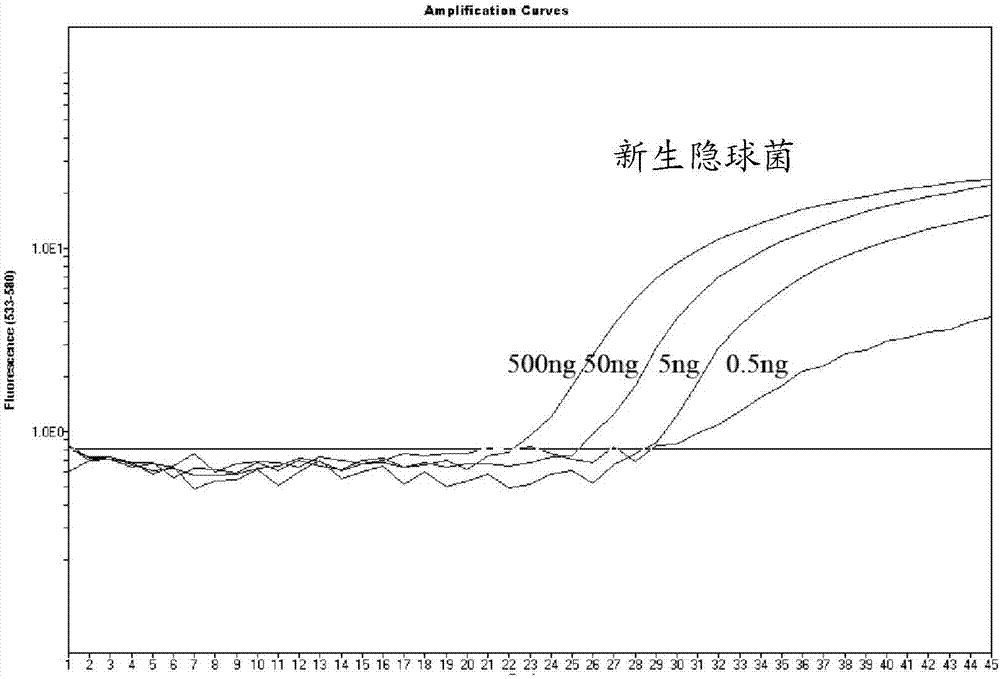
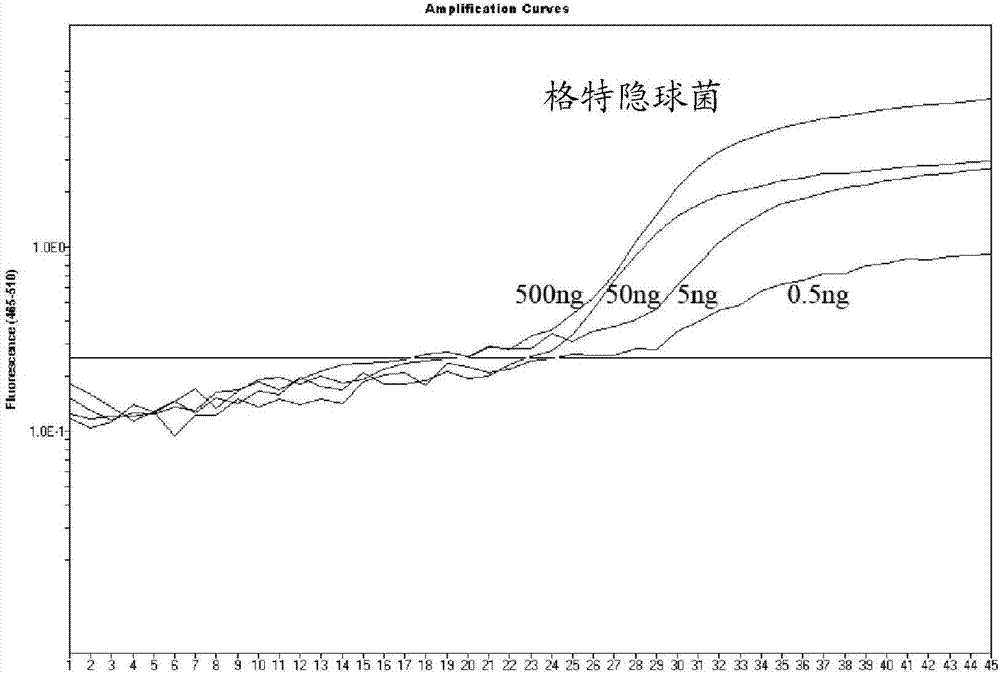
[0039] The inventors of the present invention selected the sequence fragment of the copper-zinc superoxide dismutase site as the target gene position by comparing the sequence differences between different genotypes of Cryptococcus neoformans and Cryptococcus gattii and with other fungal genes. Fragments are unique to Cryptococcus and differ between C. neoformans and C. gattii. The obtained nucleotide sequence for detecting the target gene of Cryptococcus neoformans and Cryptococcus gattii is shown in SEQ ID No.1 and / or SEQ ID No.2, wherein SEQ ID No.1 corresponds to Cryptococcus neoformans, and SEQ ID No.2 corresponds to Cryptococcus gattii.
[0040] Step 2: Design of Primers and Probes
[0041] Primer design software was used to design primers for the selected target gene posit...
Embodiment 2
[0071] Example 2: Primers, probes and kits for the detection and identification of Cryptococcus neoformans genes
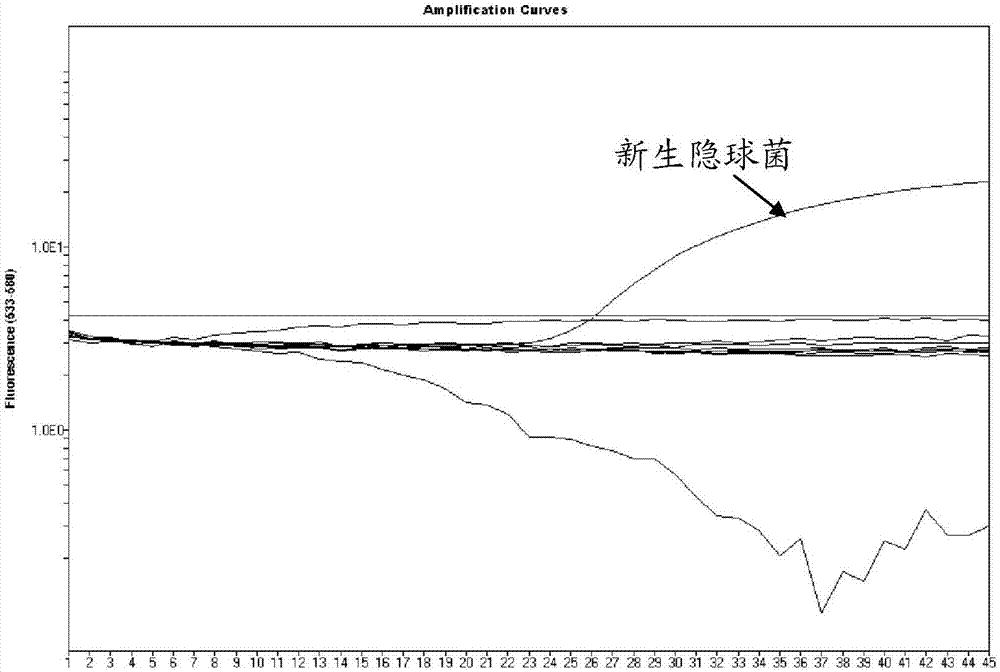
[0072] Change the primers (CN2F, CN2R, CG3F, CG3R) of detecting Cryptococcus neoformans and Cryptococcus gattii to the primers CN2F and CN2R for detecting Cryptococcus neoformans in step 3 in Example 1, and detect the primers of Cryptococcus neoformans and Cryptococcus gattii The probes (CN2P, CG2P) were changed to the probe CN2P for detecting Cryptococcus neoformans, and the remaining steps of the experiment were completed. The designed primers specifically amplified Cryptococcus neoformans, and the designed probes could further identify Cryptococcus neoformans.
Embodiment 3
[0073] Example 3: Primers, probes and kits for the detection and identification of Cryptococcus gattii genes
[0074] Change the primers (CN2F, CN2R, CG3F, CG3R) of detecting Cryptococcus neoformans and Cryptococcus gattii in step 3 in Example 1 to the primers CG3F and CG3R for detecting Cryptococcus gattii, and detect Cryptococcus neoformans and Cryptococcus gattii The probes (CN2P, CG2P) of Cryptococcus gattii were changed to the probe CG2P for detecting Cryptococcus gattii, and the remaining steps of the experiment were completed. The designed primers specifically amplified Cryptococcus gattii, and the designed probes could further identify the Teptococcus.
[0075] Use primer and probe of the present invention to carry out pathogenic bacteria detection to clinical infection patient, clinical case verification is the following table:
[0076] Clinical data and strain identification results of patients infected with Cryptococcus neoformans and Cryptococcus gattii
[0077] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com