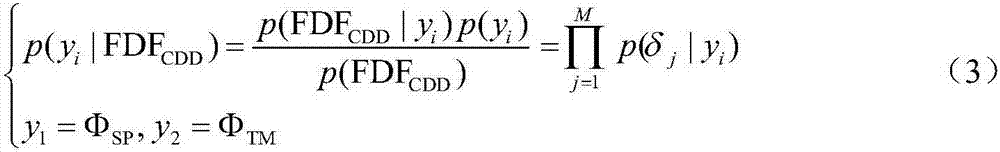Prediction method for signal peptide and cleavage site thereof on the basis of layered mixture model
A cleavage site and hybrid model technology, applied in informatics, sequence analysis, bioinformatics, etc., can solve the problems of high false positive signal peptides, and it is difficult for classifiers to correctly identify signal peptides and transmembrane helices, so as to reduce false positives. Positive, performance-enhancing, sensitivity-enhancing effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
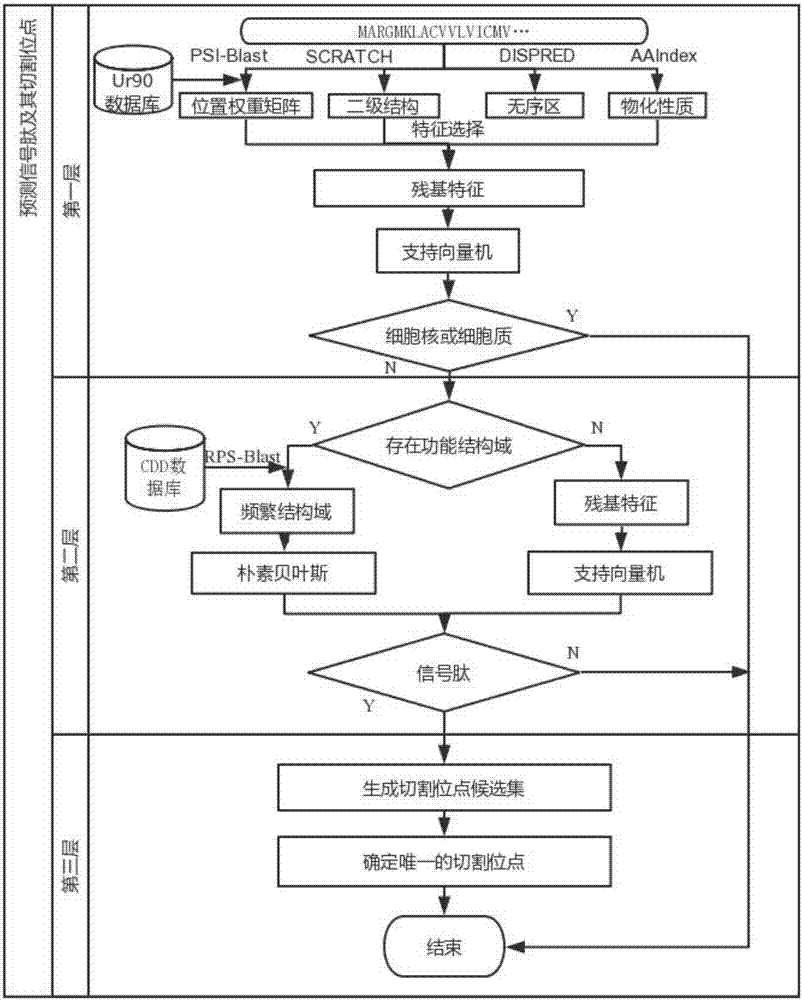
Method used
Image
Examples
Embodiment
[0043] There is an input sequence with the following data:
[0044] >QuerySequence|SIGNAL 1 25
[0045] MIKSNRITACALAALFAGASFSASAWWGGPGYGNGLWDNMGDMFGDGYGDFNMSM
[0046] GGGGRGYGRGYGRGNGYGYGAPYGYGAPYGYGAPYGYGAPYGYGAPYGAMPYGA
[0047] MPPQMPAAPAQPQAAPSR
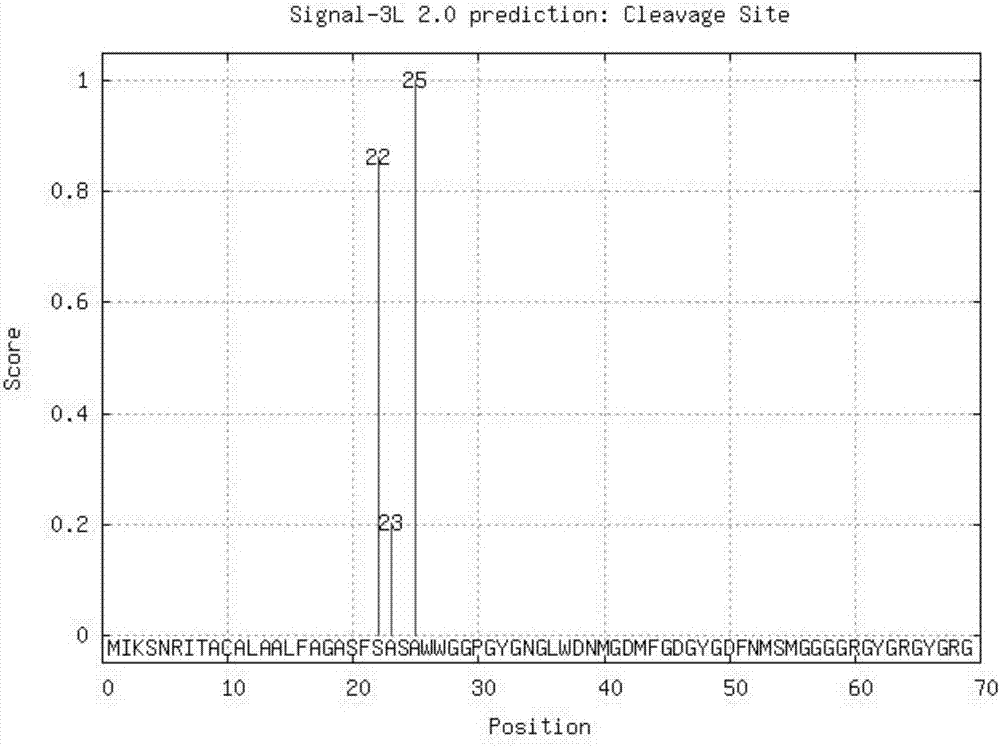
[0048] This is a sequence to be tested, and the software output results using the method of the present invention are as follows: figure 2 Shown:
[0049] According to Signal-3L 2.0 engine, the predicted signal peptide is: 1-25
[0050] MIKSNRITACALAALFAGASFSASAWWGGPGYGNGLWDNMGDMFGDGYGDFNMSM
[0051] GGGGR GYGRGYGRGN
[0052] The potential cleavage sites and the credit scores
[0053] It can be seen from the results that the method accurately and intuitively predicts the signal peptide and its cleavage site.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com