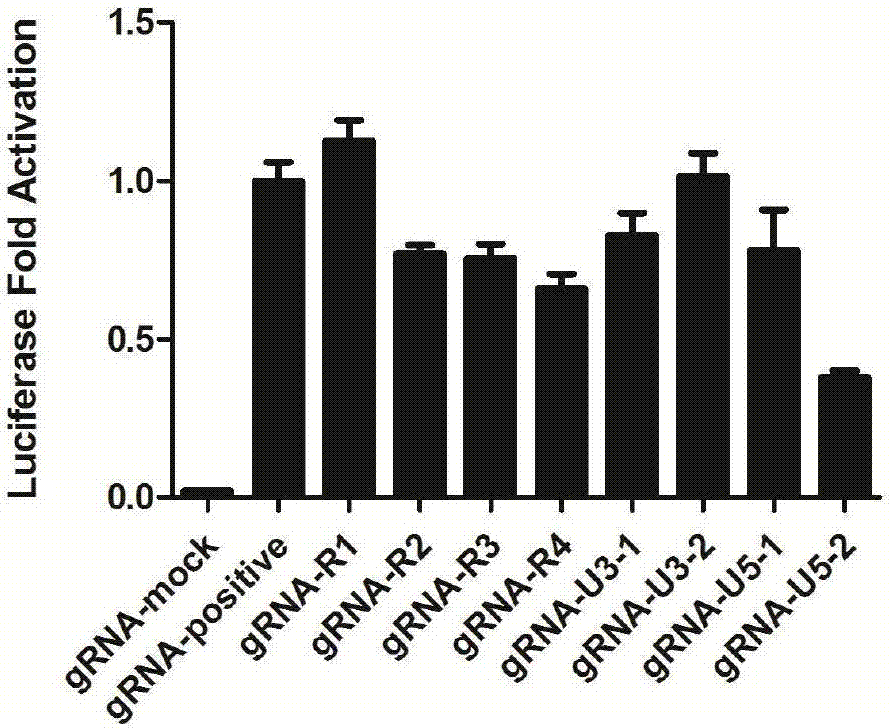
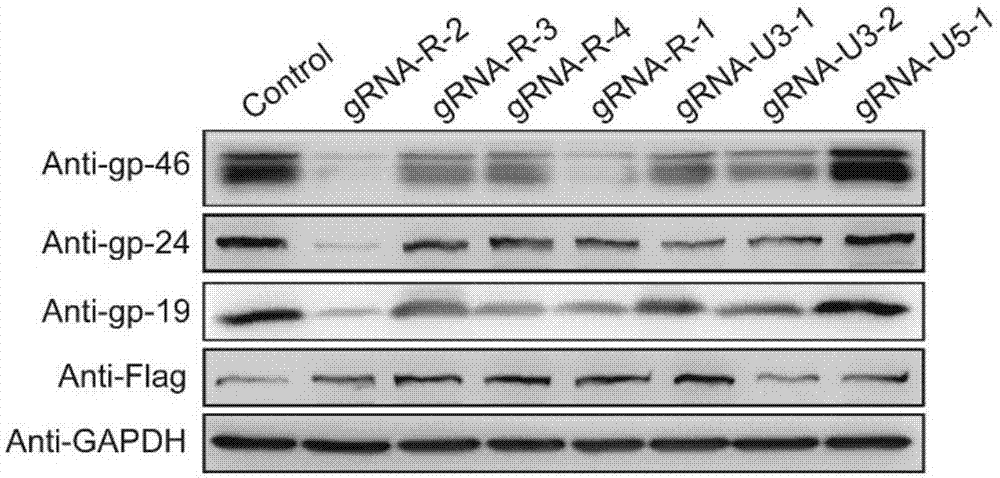
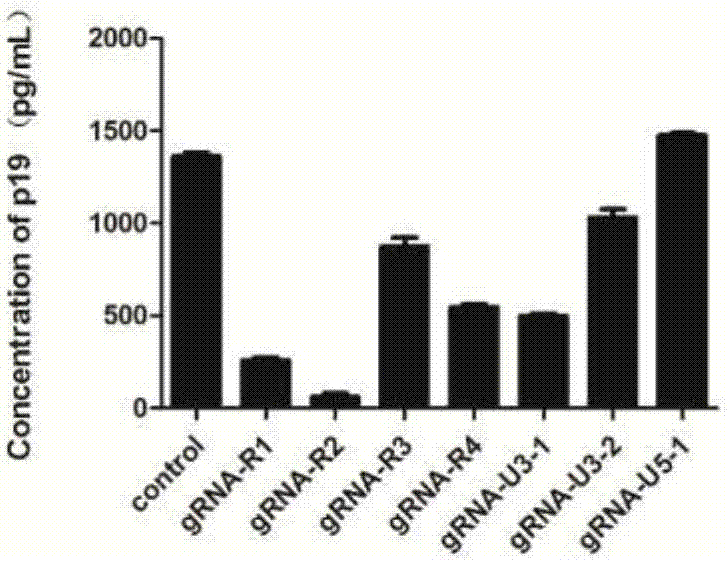
GRNA (guide Ribonucleic Acid) sequence capable of effectively knocking out CRISPR/Cas9 (Clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9) of HTLV-1 (Human T-cell Leukemia Virus type 1) virus genome
A virus genome and sequence technology, applied in the field of gRNA sequences, can solve the problems of secondary infection, ineffective removal of HTLV-1, etc., and achieve the effect of inhibiting proliferation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Recombinant plasmid primer design and target selection for constructing targeting sequence
[0027] Taking the LTR region of HTLV-1 as the target selection region, in this region, based on the basic principles of primer design, design several pairs of upstream and downstream primer sequences for different targets:
[0028] R1:
[0029] DNA forward sequence: CACCGGACTCAACCGGCGTGGATGG (SEQ ID NO 05, which is the first DNA forward sequence of the present invention, corresponds to the first RNA forward sequence, as shown in SEQ ID NO 01:
[0030] GACUCAACCGGCGUGGAUGG)
[0031] DNA reverse sequence: AAACCCATCCACGCCGGTTGAGTCC (SEQ ID NO 06, which is the first DNA reverse sequence of the present invention, corresponds to the first RNA reverse sequence, as shown in SEQ ID NO 02:
[0032] CCAUCCACGCCGGUUGAGUC)
[0033] R2:
[0034] DNA forward sequence: CACCGAGAACGCGACTCAACCGGCG (SEQ ID NO 07, which is the second DNA forward sequence of the present invention, corresponds to the se...
Embodiment 2
[0097] Example 2: Detection of various target indicators in cells
[0098] The plasmids constructed in Example 1 were packaged with viruses and infected cells, and the cells were tested for various target indicators
[0099] 1. Lentivirus packaging and stable strain screening
[0100] A. Lentivirus packaging
[0101] (1) Collect 293FT cells the day before transfection to 2×10 per plate 6 Cells (10mL cell suspension) were inoculated into a 10cm petri dish, 37℃, 5% CO 2 to cultivate. (293FT medium contains: DMEM, 10% FBS, P / S);
[0102] (2) After 12 hours, add 1mL OPTI-MEM to a 1.5mL EP tube, add 75μL Lipofectamine3000, and mix upside down; take another 1.5mLE tube and add 1mL OPTI-MEM and prepare the plasmid according to the following system: LentiCRISPRv2-gRNA (15μg ), pCMV-A8 / 9(15μg), pcDNA-VSVG(7.5μg)
[0103] (3) Mix upside down;
[0104] (4) Mix the above two mixtures together, let them stand at room temperature for 5 minutes, and drop them evenly on a 10 cm culture plate;
[0105] (...
Embodiment 3
[0237] Example 3 Animal experiment
[0238] The stable strain obtained in Example 2 was injected into mice to construct a mouse tumor model
[0239] (1) Collect cells and count them
[0240] (2) The left and right sides of each mouse's back were injected with the cells of the control group and the experimental group, the number was 7×10 5 Pcs / only
[0241] (3) After 4 weeks, the mice were dissected, tumors were removed, weighed, and various indicators of tumor cells were detected
[0242] The result is Figure 7 As shown in (A-C), both the tumor formation rate and the tumor volume of the experimental group were significantly lower than those of the control group; in addition, we tested the tumor-forming body of each group of mice through immunohistochemistry experimental technology, and the results are as follows Figure 7 As shown in (D), tumors appeared in the experimental group and cells in the control group not only formed larger tumors but also grew in good tumor cells.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com