Oligonucleotide library for specific identification of whole short arm of ninth chromosome of cultivated rice and specific identification method
An oligonucleotide library and specific recognition technology, which is applied in the field of specific recognition of the whole short arm oligonucleotide library of the No. 9 chromosome of cultivated rice and can solve the problem of no specific recognition
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
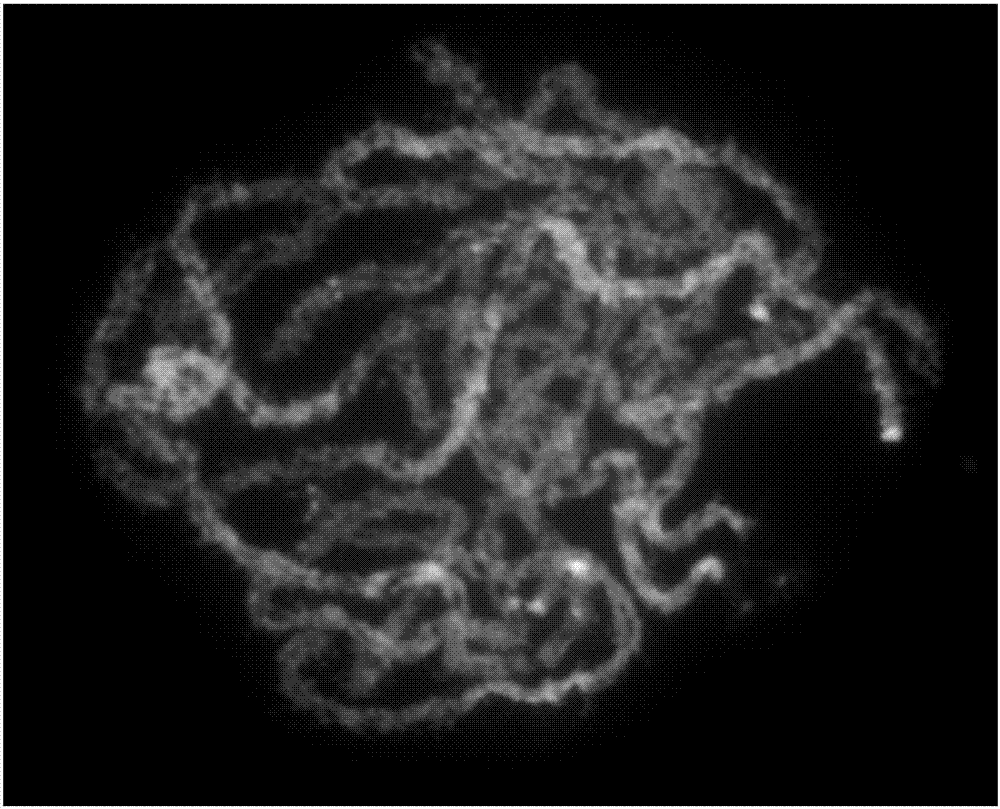
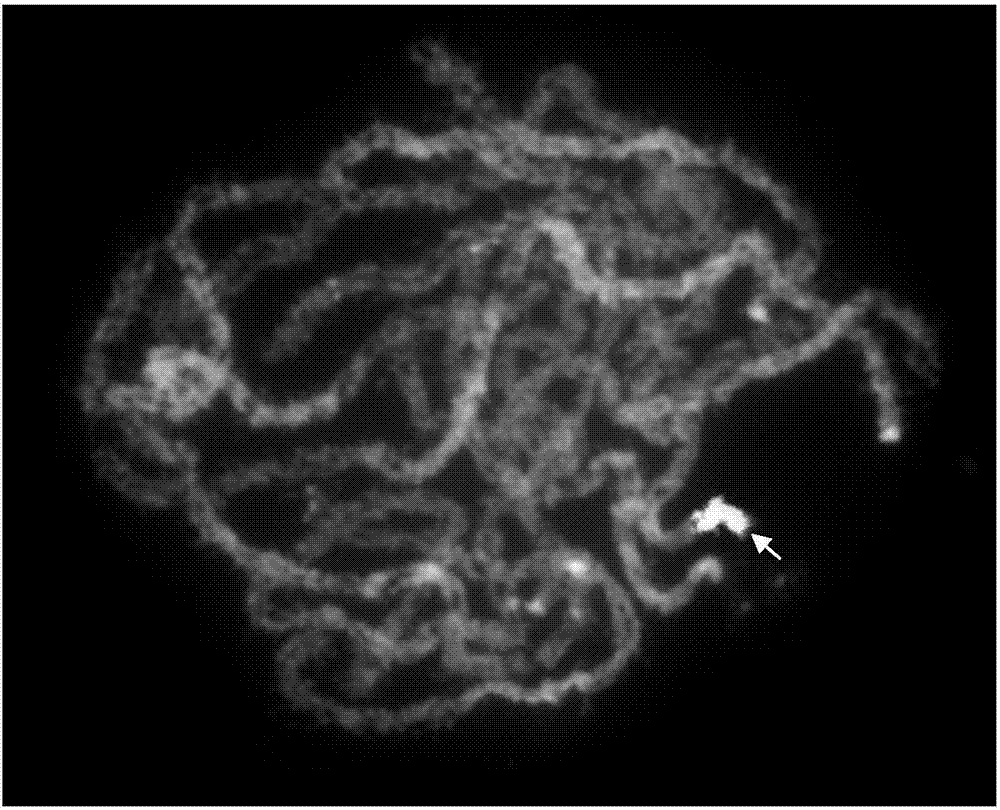
[0018] 1. Using bioinformatics capture technology to screen 2258 oligonucleotides distributed in the entire short arm of Nipponbare chromosome 9:
[0019] Step 1: Screen and exclude all repetitive sequences in the entire short arm of chromosome 9 of the cultivated rice Nipponbare.
[0020]The second step: oligonucleotide screening: use Chorus software (https: / / github.com / forrestzhang / Chorus), (design parameters are: oligonucleotide length is 45nt, homology parameter is 75, dTm>10° (dTm=Tm-hairpinTm) oligonucleotides can be used for library construction (Tm: annealing temperature of oligonucleotides, hairpin Tm: hairpin temperature) to screen 2258 specific oligonucleotides distributed on the short arm of Nipponbare chromosome 9 Nucleotide, the sequence is shown in SEQ ID NO.1-2258.
[0021] 2. Synthesize an oligonucleotide library (2258 oligonucleotides) for probe labeling (completed by MYcroarray Company, Ann Arbor, MI). 3. Preparation of single-stranded oligonucleotide prob...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com