Preparation method of endophytic bacterial diversity 16SrDNA amplicon
A plant endophytic bacteria and amplicon technology, applied in the field of high-throughput sequencing of microbial diversity, can solve the problem of high proportion of host chloroplast data, and achieve the effect of reducing the proportion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
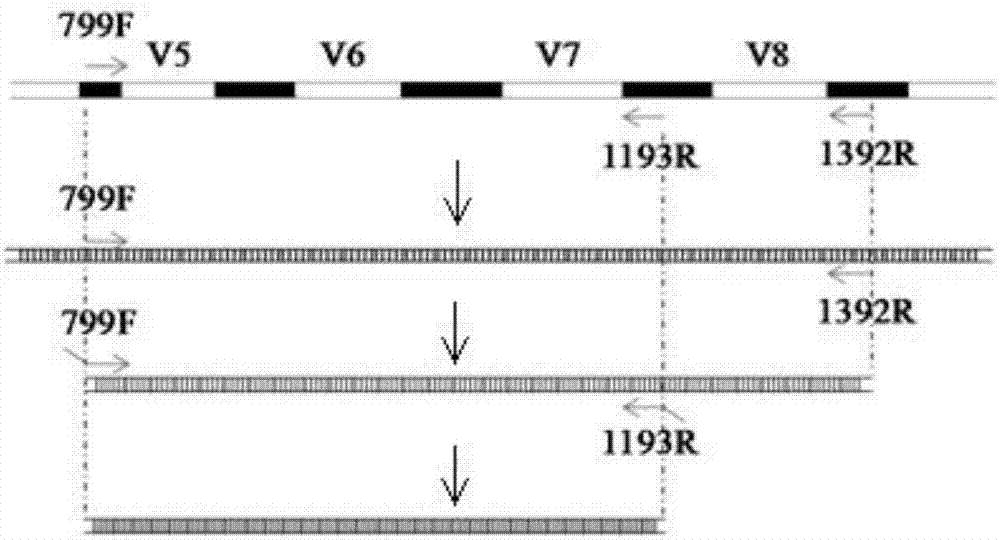
[0063] This example provides a set of primers for high-throughput sequencing. The primers consist of the primer pair 799F / 1193R for the 16S rDNA V5-V7 variable region fragment of plant endophytic bacteria and the 5 primers connected to the primer pair 799F / 1193R. 'end Barcode sequence composition, wherein, the sequence of the 799F is SEQ ID NO: 1, the sequence of the 1193R is SEQ ID NO: 4, and the Barcode sequence connected to the 5' end of the 799F is selected from SEQ ID NO: For the sequences in 5-28, the Barcode sequence connected to the 5' end of the 1193R is selected from the sequences in SEQ ID NO: 29-52.
[0064] Specifically, the primer sequences and names involved in the present invention are summarized as follows:
[0065]
[0066]
[0067]
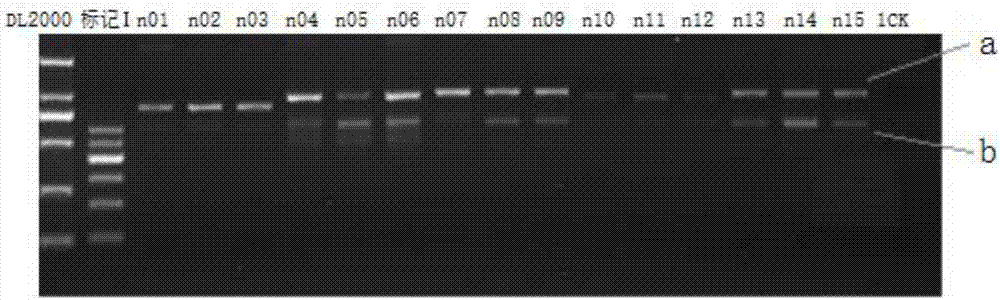
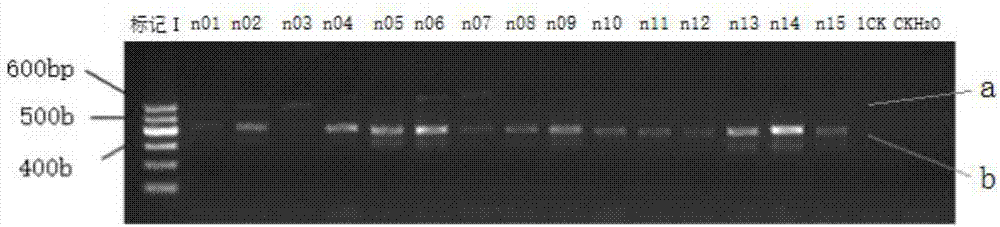
[0068] Further, the embodiment of the present invention provides a method of using the above-mentioned primer and primer pair 799F / 1392R to prepare a plant endophytic bacterial diversity 16S rDNA amplicon suitable for a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com