Method for utilizing machine learning to predict complex disease susceptibility locus
A machine learning and disease technology, applied in the fields of instrumentation, informatics, genomics, etc., to achieve the effect of improving heritability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0030] The content of the present invention will be described in further detail below in conjunction with the accompanying drawings.
[0031] Example: Taking the complex disease type II diabetes as an example, the method of the present invention is used to predict the susceptibility loci of type II diabetes, which will be described in detail below.
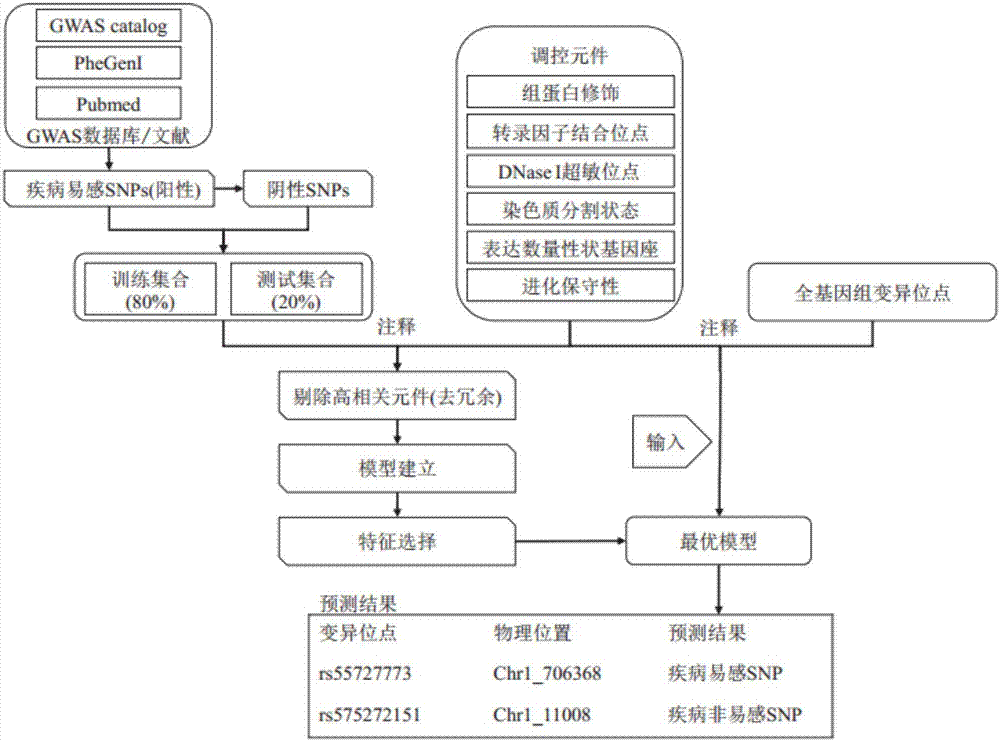
[0032] Such as figure 1 As shown, the present invention provides a screening method for predicting complex disease susceptibility loci based on the characteristics of genome epigenetic regulatory elements by using machine learning, including the following steps P1-P3.
[0033] P1: Collect known type 2 diabetes susceptibility loci as positive sets for machine learning models and perform annotation of epigenetic regulatory elements.
[0034] It specifically includes: collecting 65 known susceptibility SNPs for type 2 diabetes from relevant literature in the public database GWAS catalog, PheGenI and Pubmed, as a positive set. After...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com