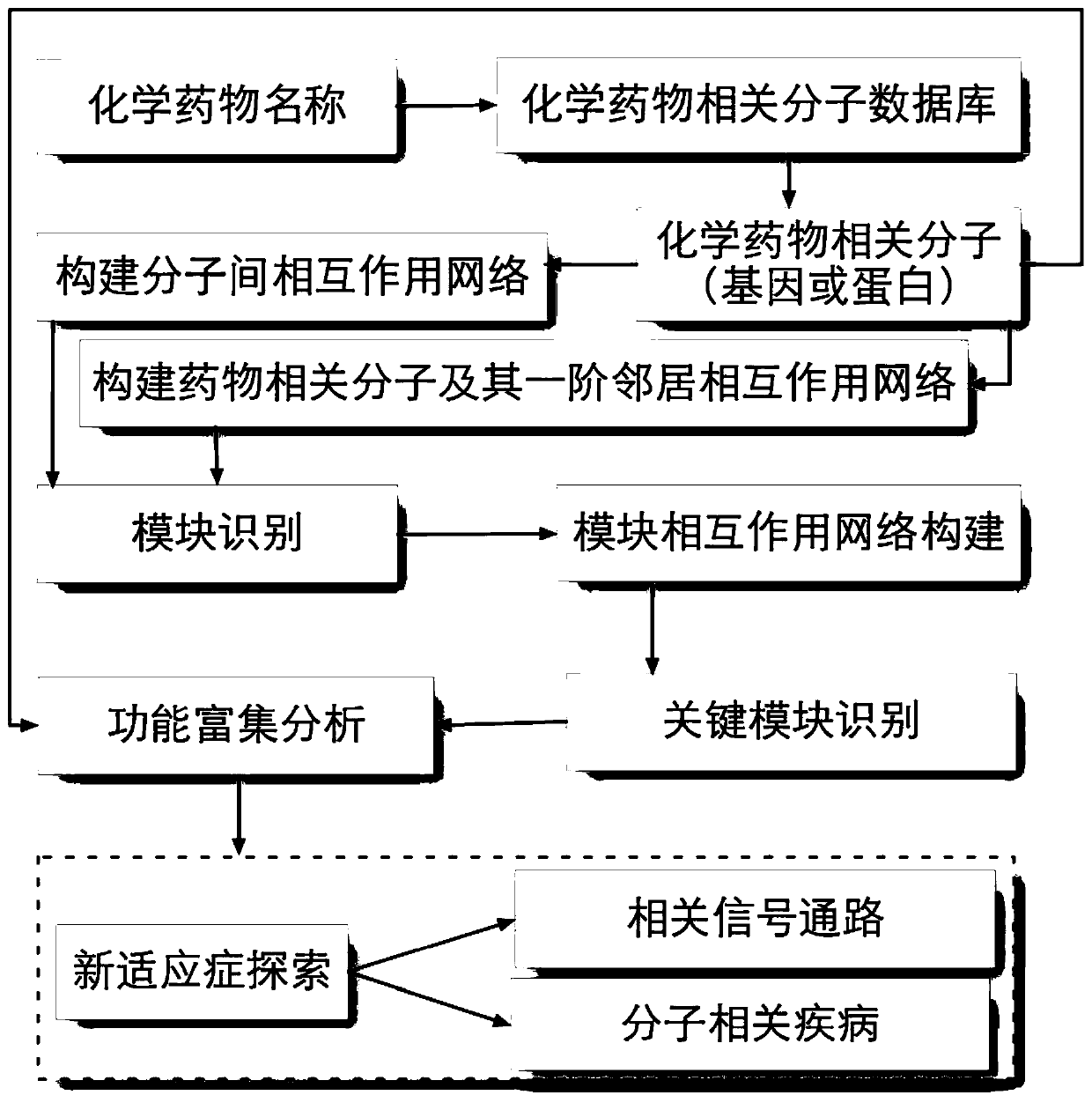
A way to explore new indications for drugs
A new indication and drug technology, applied in the field of bioinformatics, can solve problems such as low success rate, negative impact of development projects, high cost investment, etc., and achieve the effect of saving manpower, reducing research and development risks, and rational use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1 On the basis of Sorafenib's existing commonly used treatments for renal cell carcinoma and hepatocellular carcinoma, explore its new indications through network pharmacology methods
[0060] Enter the name of the drug Sorafenib into the CTD (Comparative Toxicogenomics Database-http: / / ctdbase.org / ) database to find the related genes. A total of 148 were found, see Table 2.
[0061] Table 2. Sorafenib-associated genes found in CTD
[0062]
[0063]
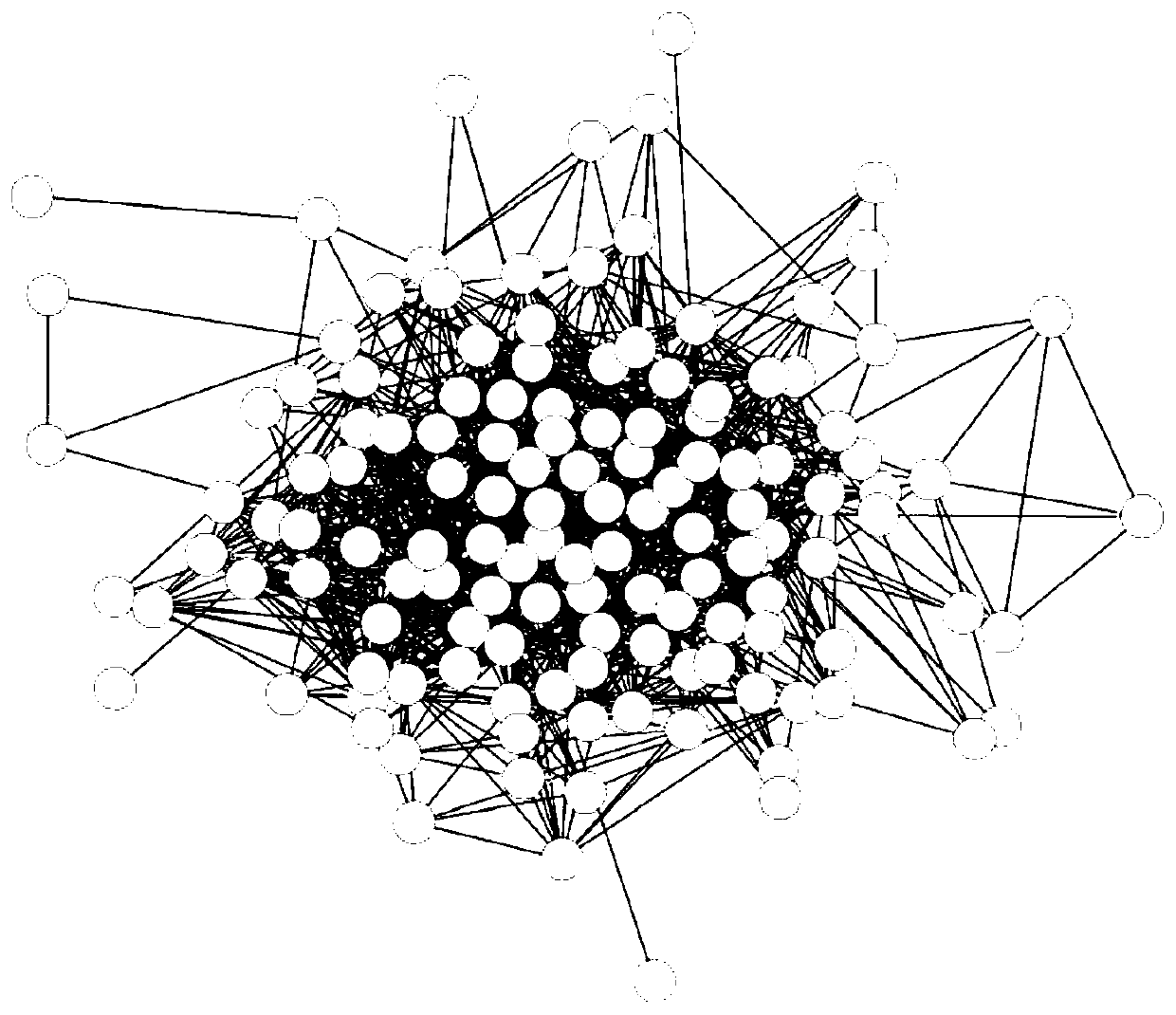
[0064] After the genes are mapped and extracted from the protein-protein interactions of related genes through the STRING database platform, the protein-protein interaction network of Sorafenib-related proteins is obtained, which contains 133 nodes and 1040 edges ( figure 2 ).
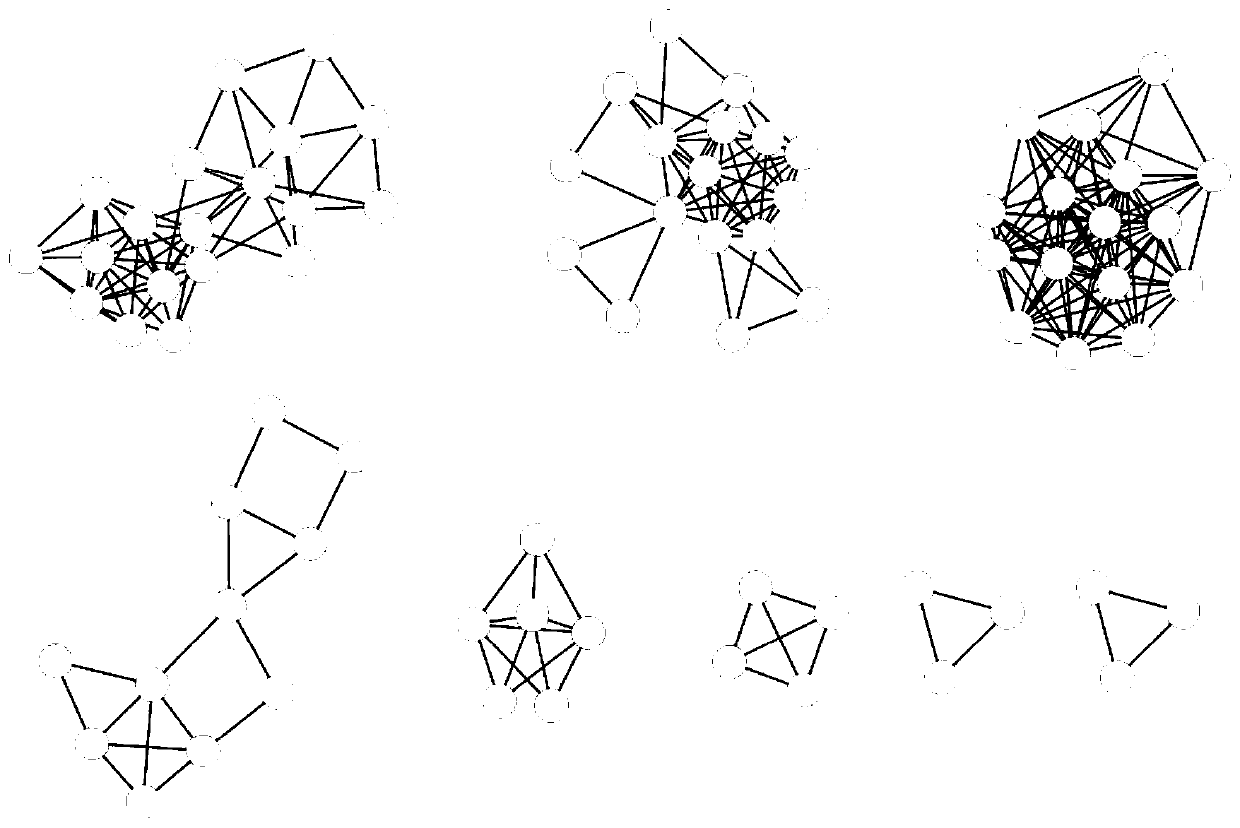
[0065] The MCODE method (cytoscape3.0 version) was used to identify the modules of the interaction network, and the parameters were set as Degree Cutoff: 2; Node Score Cutoff: 0.2; K-Core: 2; Max Depth: 100. 8 modules are obtaine...
Embodiment 2
[0082] Example 2 On the basis of the existing commonly used oral hypoglycemic agents of acarbose (Acarbose), its new indications were explored through the network pharmacology method.
[0083] Enter the drug Acarbose (Acarbose) name into the CTD (Comparative Toxicogenomics Database-http: / / ctdbase.org / ) database to find the related genes. A total of 14 related genes were obtained, as shown in Table 6.
[0084] Table 6. Acarbose-related genes found in CTD
[0085]
[0086] After the gene is mapped by the STRING database platform, the protein of the related protein and its first-order neighbor interaction are extracted, and the interaction network of the protein of the acarbose-related protein and its first-order neighbor is obtained, which contains 507 nodes and 5911 edges ( Image 6 ), where the black nodes are acarbose-related proteins, and the white nodes are protein-related first-order neighbors.
[0087] The MCODE method (cytoscape3.0 version) was used to identify th...
Embodiment 3
[0106] Example 3 On the basis of axitinib (axitinib), which is commonly used in the treatment of kidney cancer, its new indications are explored through network pharmacology methods.
[0107] Enter the name of the drug axitinib into the CTD (Comparative Toxicogenomics Database-http: / / ctdbase.org / ) database to search for related genes. A total of 7 were found, see Table 11.
[0108] Table 11. Axitinib-related genes found in CTD
[0109] Axitinib Gene VEGFA FLT1 FLT4 KDR NOS3 PAK1 THBS1
[0110] After the genes are mapped and extracted from the protein-protein interactions of related proteins by the STRING database platform, the protein-protein interaction network of axitinib-related proteins is obtained, which contains 7 nodes and 17 edges ( Figure 10 ).
[0111] DAVID software was used to perform functional enrichment analysis of the interaction network from the perspective of KEGG pathways, and the results of P<0.01 were retained, and a total of 7...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com