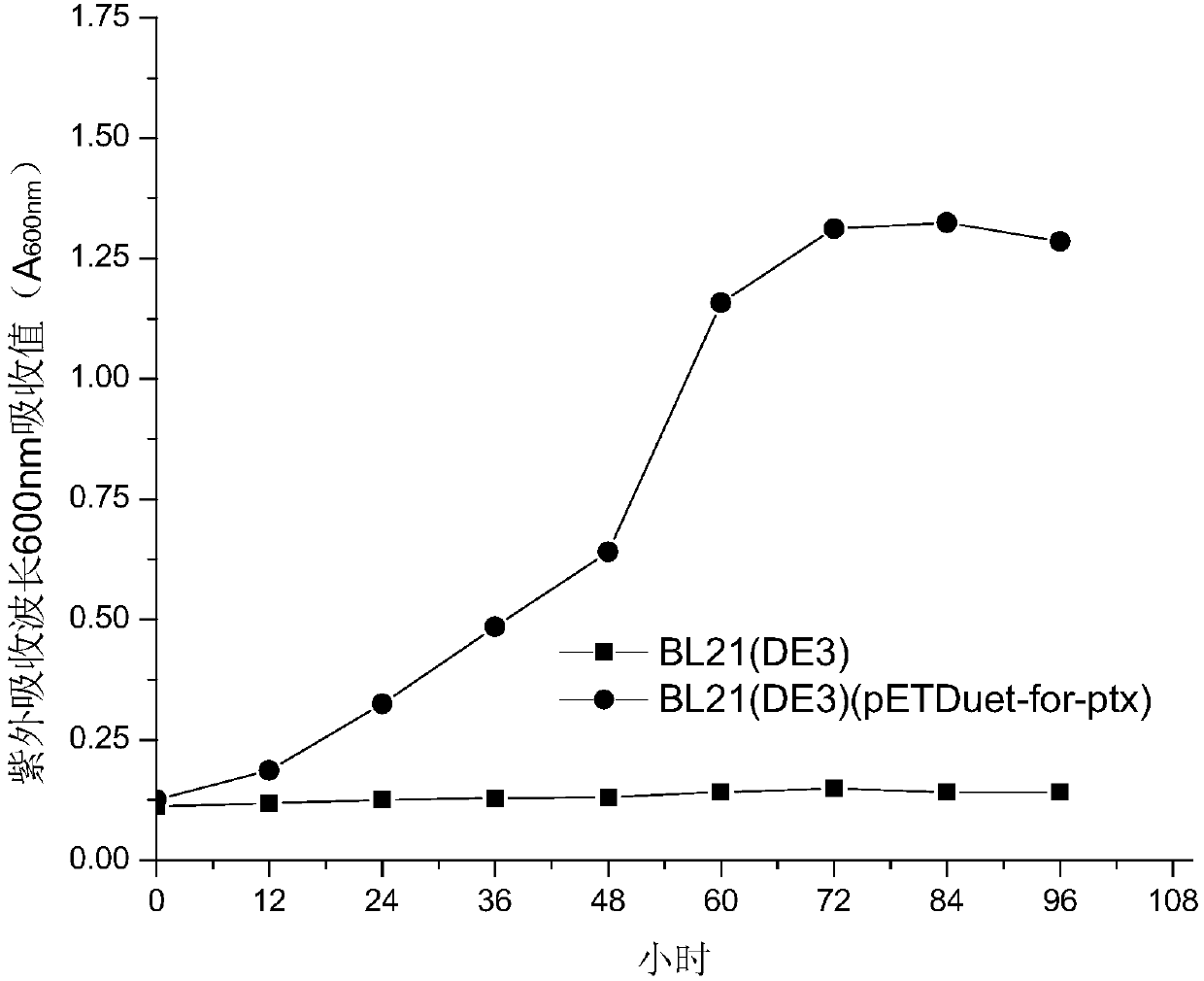
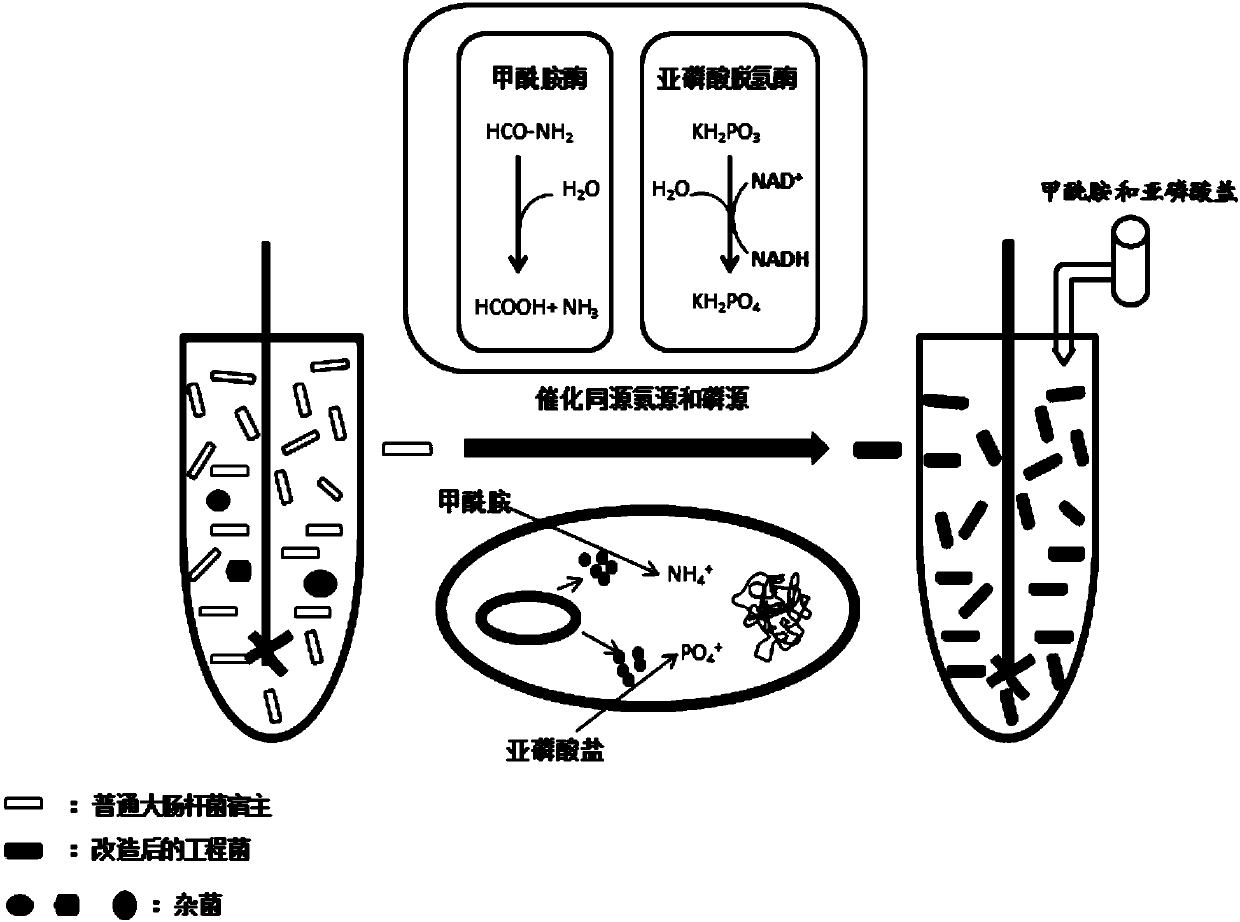
Recombinant escherichia coli capable of efficiently decomposing formamide and phosphate oxide, and construction method and application thereof
A technology of recombining Escherichia coli and oxidizing phosphite, which is applied in the field of genetic engineering, can solve the problem of "killing" and achieve the effects of strong purpose, no contamination of bacteria products, and high material purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
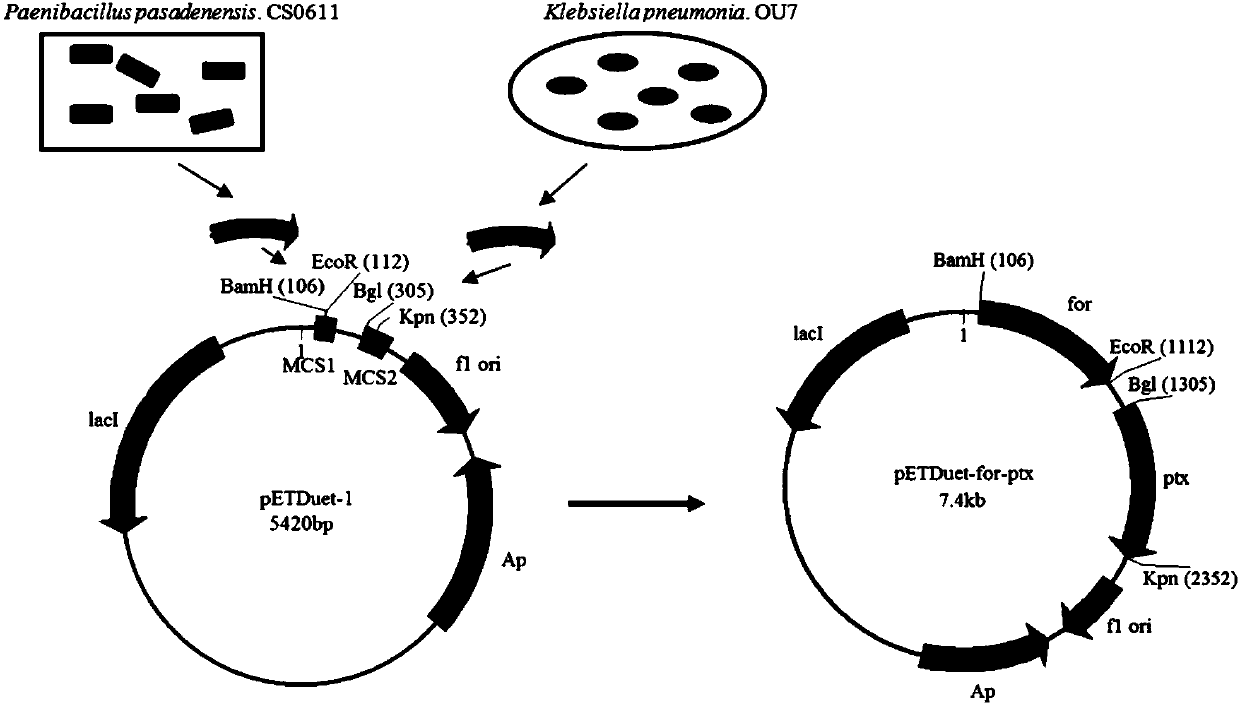
[0054] Acquisition of formamidase gene for
[0055] Cultivate Paenibacillus pasadenensis.CS0611 with LB medium for one day at 37°C and 180rpm; collect the cultured cells by centrifuging at 4°C and 8000rpm for 5 minutes, and wash them twice with normal saline to remove residual culture base, and then extract the Paenibacillus paenibacillus pasadenensis.CS0611 genome according to the specific method of the OMEGA bacterial genome DNA extraction kit;
[0056] Using the extracted Paenibacillus pasadenensis.CS0611 genome as a template, A1(5'-CGC GATGAACGGACTGGGCGGCTTGAAC-3') and A2(5'-CCG CGTCGCGCCGCGCCTCCCTTCGCTC-3') are the upstream and downstream primers respectively, and the formamidase gene is amplified by PCR;
[0057] The enzyme reagent used in PCR is TaKaRa company's HS DNA Polymerase with GCbuffer; PCR reaction system and conditions are as follows:
[0058]
[0059]
[0060] PCR reaction conditions: react at 94°C for 5min; then react at 98°C for 10s, 55°C for 5...
Embodiment 2
[0063] Acquisition of phosphite dehydrogenase gene ptx
[0064] The phosphite dehydrogenase gene was derived from Klebsiella pneumonia.OU7, which was independently screened, and was obtained through independent screening. The screening medium used Monkina solid medium (g): glucose 10, (NH 4 ) 2 SO 4 0.5, MgSO 4 ·7H 2 O 0.3, NaCl 0.3, KCl 0.3, FeSO 4 ·7H 2 O 0.0036, MnSO4 ·H 2 O 0.03, KH 2 PO 3 0.3, Agar 20, dilute to 1L with ultrapure water;
[0065] Dip the single colony grown on the screening plate with an inoculation needle, and then continue to grade and streak on a new screening plate until a single colony appears to achieve purification, and finally isolate and purify a strain that grows on this screening medium A very good strain, through 16S rDNA sequencing, showed that it was Klebsiella pneumonia.OU7.
[0066] According to the specific method of the OMEGA Bacteria Genomic DNA Extraction Kit, the genome of Klebsiella pneumonia. -GA GATGCTGCCGAAACTCGTTATA...
Embodiment 3
[0072] Construction of Recombinant Bacteria BL21(DE3)(pETDuet-for)
[0073] The gene fragment for and plasmid pETDuet-1 obtained in Example 1 were double-digested with BamH I and EcoR I respectively. The underlined italics of the upstream primers were the BamH I enzyme cut points, and the underlined italics of the downstream primers were the EcoR I enzyme cut points. Cutting conditions: 37°C, enzyme digestion for 60 minutes;
[0074]
[0075] The digested products were recovered by gel cutting, and the recovery method and steps refer to the gel recovery kit of OMEGA Company; after recovery, 1wt% agarose gel electrophoresis was used to detect the recovery rate; then the recovered target fragment was connected to the plasmid, The ligation kit uses T4DNA Ligase from Thermo Fisher SCIENTIFIC, and the ligation system is as follows:
[0076]
[0077]
[0078] The molar ratio of target fragment to plasmid is 3:1.
[0079] Transform the ligation product into Escherichia col...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap