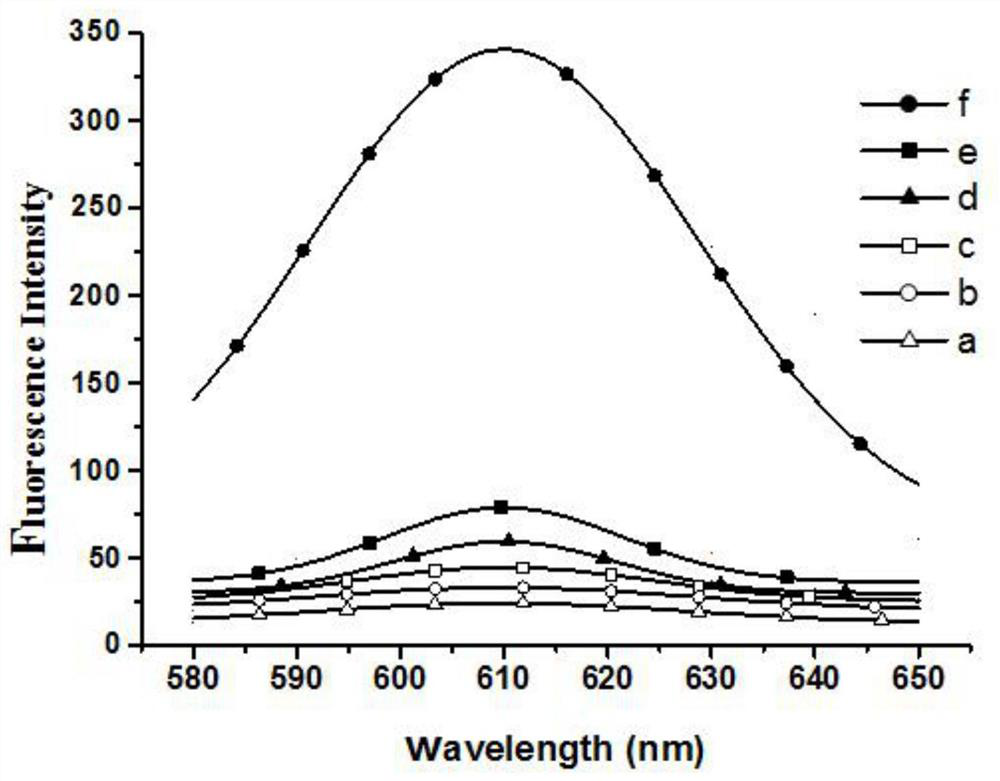
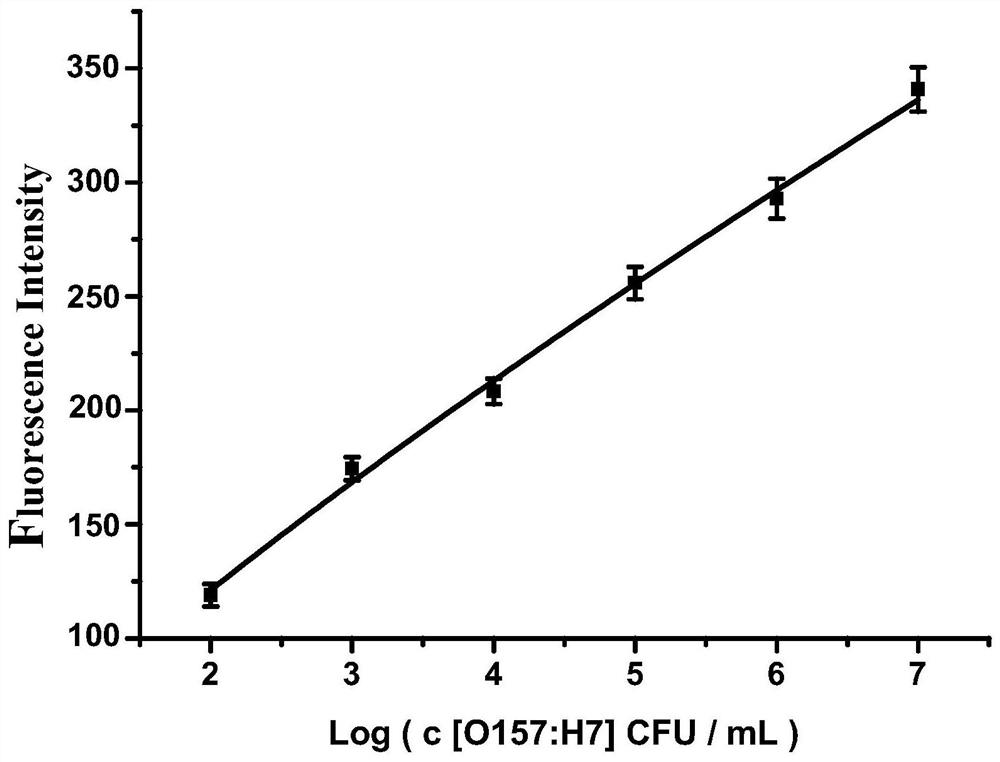
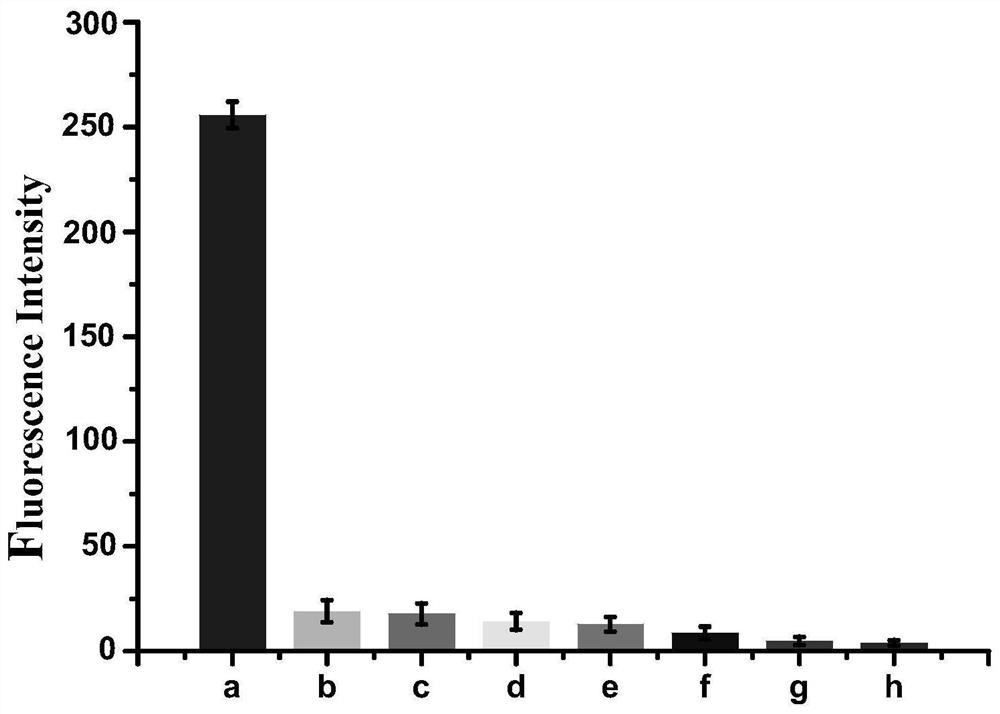
Detection method of e.coli O157:H7 free from enzyme and fluorescence labeling
A detection method, fluorescence intensity technology, applied in fluorescence/phosphorescence, measurement device, material analysis by optical means, etc., to achieve a highly specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] (1) Hybridize the IS with the aptamer (conditions for hybridization: at 80-90°C, treat for 40-55min), the nucleotide sequence of the IS is shown in SEQ ID NO: 1, and the aptamer The nucleotide sequence of the gamete is shown in SEQ ID NO: 2;
[0033] (2) Place the product obtained in step (1) in a buffer, add Escherichia coli O157:H7, and react at 37°C for 30 minutes;
[0034] (3) Add GHP1 and GHP2 to the result of step (2), and incubate at 37° C. for 100 min; the nucleotide sequence of GHP1 is shown in SEQ ID NO: 3, and the nucleotide sequence of GHP2 is shown in SEQ ID NO : as shown in 4;
[0035] (4) Add NMM to the resultant of step (3), and incubate at 37° C. for 30 min; the NMM is N-methyl mesoporphyrin IX;
[0036] (5) Perform fluorescence intensity detection on the product obtained in step (4).
[0037] The IS and aptamers were treated at 90° C. for 5 minutes before use, and then cooled slowly to room temperature.
[0038] The concentration of the IS was 3 μM...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com