A kind of diblock molecular probe and its rapid detection method for nucleic acid
A molecular probe and diblock technology, which is used in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of increased detection cost, time-consuming, etc. Low cost and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0038] 1. Preparation of diblock probes
[0039] (1) Design, synthesis and purification of diblock probes:
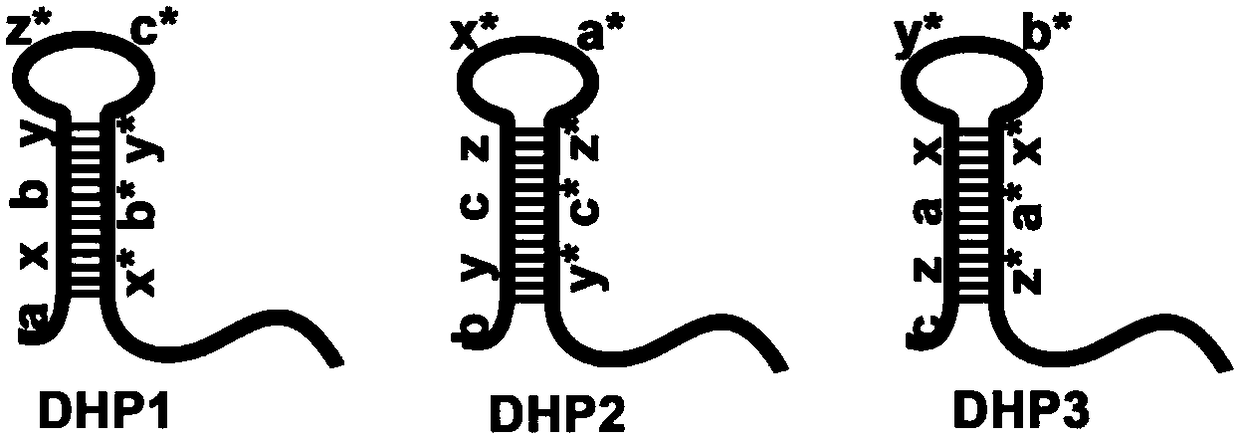
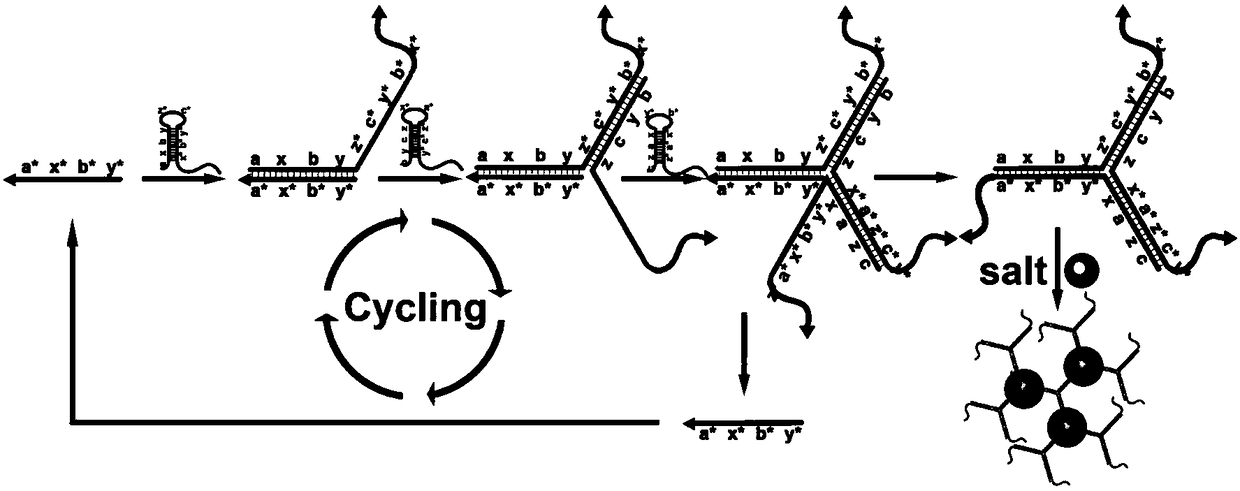
[0040] Design three diblock molecular probes: diblock molecular probe 1, diblock molecular probe 2 and diblock molecular probe 3; the diblock molecular probe 1 is sequentially from 5' to 3' Including: a promoter region, a hairpin structure region and a polyA region; wherein, the promoter region of the double-block molecular probe 1 is complementary to the partial sequence of the target DNA, and the promoter region of the double-block molecular probe 2 is complementary to the partial sequence of the double-block molecular probe The partial sequence of the hairpin structure region of 1 is complementary, the promoter region of diblock molecular probe 3 is complementary to the partial sequence of the hairpin structure region of diblock molecular probe 2, and the hairpin structure of the three diblock molecular probes After the region is opened, three double-block molecular...
Embodiment 1
[0053] 1. Preparation of diblock probes
[0054] (1) Design and synthesis of diblock probes
[0055] Design a diblock probe based on the target DNA sequence. The nucleic acid sequence of the target DNA is as follows:
[0056] y* b* x* a*
[0057] 5'-GCACTA-CTCCCT-AACATC-TCAAGC-3' (SEQ ID NO.1)
[0058] For the designed diblock probe, use NUPACK software to analyze the free energy of its two-dimensional structure. If the free energy is negative, the probe is feasible. The designed double-block probes were synthesized by a nucleic acid synthesis company, and the synthesized probes were purified by high-performance liquid phase, and mass spectrometry was used to detect whether the synthesized probes were consistent with the design. The successfully synthesized probes were freeze-dried and stored at -20°C. The specific sequence is as follows:
[0059] DHP1:
[0060] a x b y z* c* y* b* x*
[0061] 5'-GCTTGA-GATGTT-AGGGAG-TAGTGC-TCCAAT-CACAAC-GCACTA-CTCCCT-AACATC-AAAAAAAAAAA...
Embodiment 2
[0091] In order to further confirm that the method of the present invention is applicable to the bacterial nucleic acid detection process, according to the specific nucleic acid sequence of Shewanella outer membrane cytochrome c protein: 5'-TGGTCA-ATGTGT-TCTAGC-GAAGGT-3' (SEQ ID NO.5) , designed three new diblock probes. The specific sequence is as follows:
[0092] DHP1*:
[0093] 5'-ACCTTC-GCTAGA-ACACAT-TGACCA-TCCAAT-CACAAC-TGGTCA-ATGTGT-TCTAGC-AAAAAAAAAAAAAAAA-3' (SEQ ID NO. 6);
[0094] DHP2*:
[0095] 5'-ACACAT-TGACCA-GTTGTG-ATTGGA-ACACAT-GAAGGT-TCCAAT-CACAAC-TGGTCA-AAAAAAAAAAAAAAAA-3' (SEQ ID NO. 7);
[0096] DHP3*:
[0097]5'-GTTGTG-ATTGGA-ACCTTC-ATGTGT-TGGTCA-ATGTGT-TCTAGC-GAAGGT-TCCAAT-AAAAAAAAAAAAAAAA-3' (SEQ ID NO. 8).
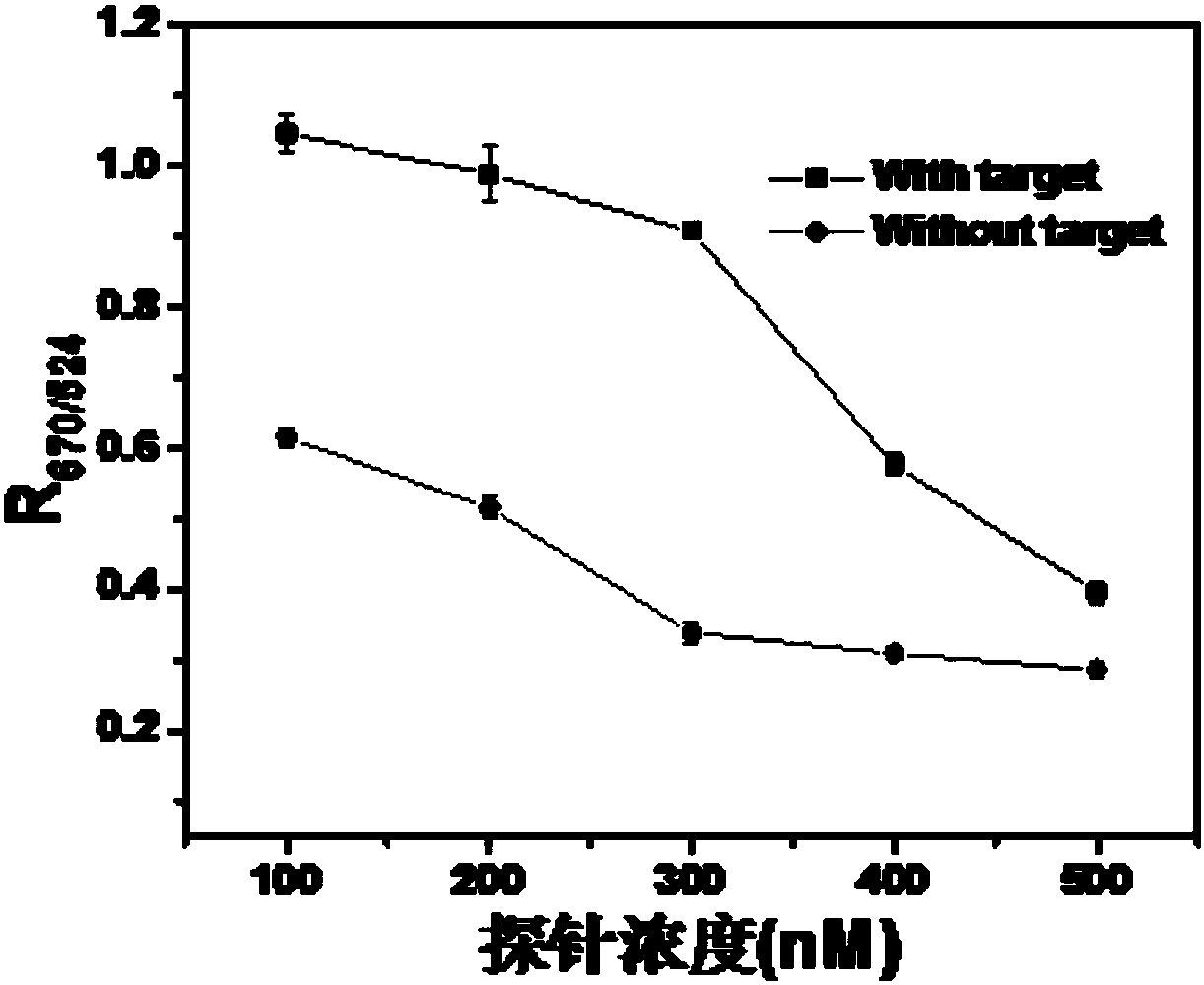
[0098] The test results show (such as Figure 8 shown), when there is a bacteria-specific probe sequence, the color change is significant, which illustrates the universality of the design of the double-block molecular probe and its rapid detec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com