Sequence-specific nuclear acid binding protein based nucleic acid detecting and ribotyping method and application thereof
A protein-binding and specific technology, applied in the field of biomedicine, can solve problems such as underutilization of value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1 Detection of HPV18 L1 gene fragments by PABE method
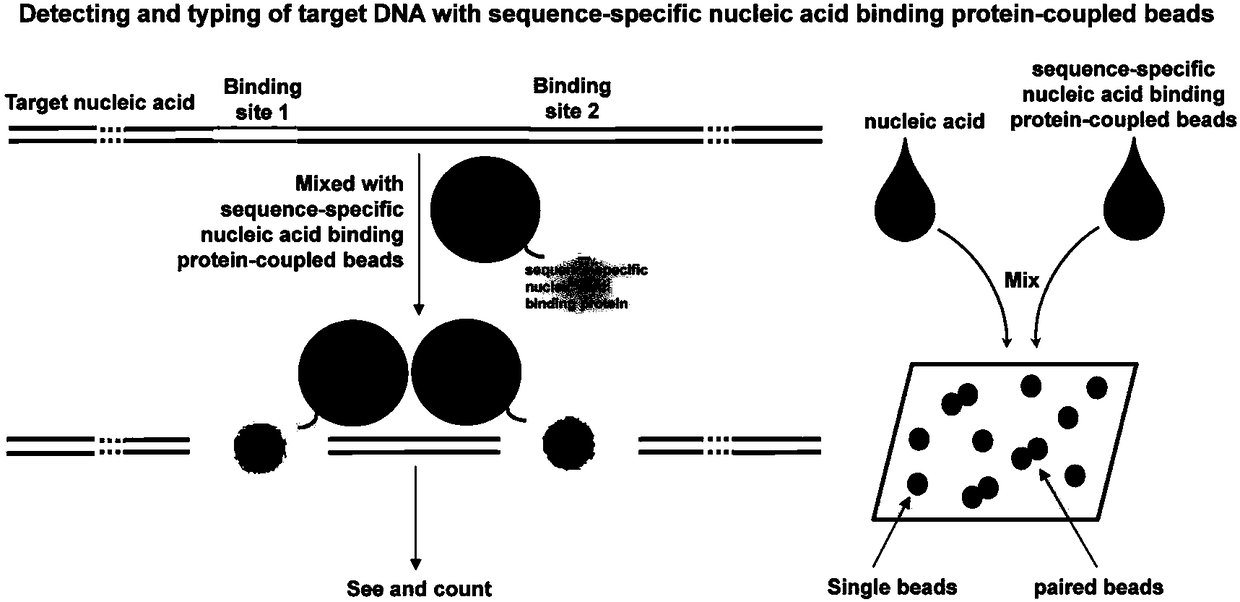
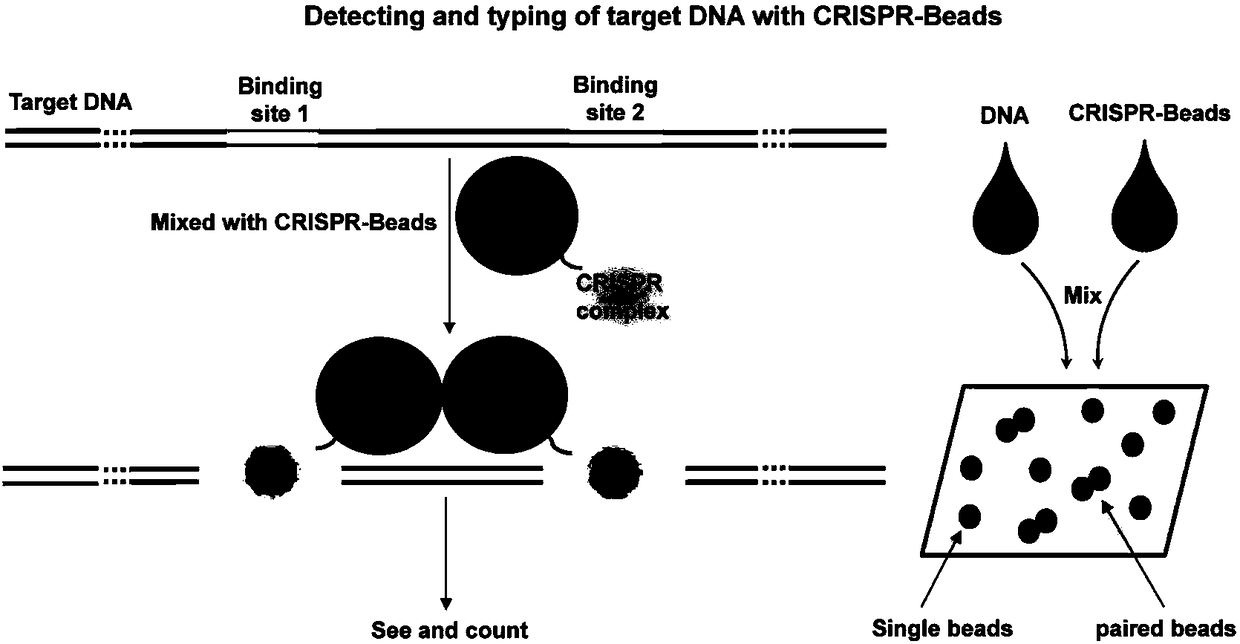
[0068] CRISPR nuclease complex Cas9 / sgRNA is a sequence-specific DNA or RNA binding protein currently used for targeted binding to a specific nucleic acid sequence, so it can be used in the PABE detection of the present invention. The detection principle is as follows: figure 2 shown. In this example 1 and examples 2-7, the 3' end of the sgRNA is added with a sequence that can anneal and hybridize with the capture sequence immobilized on the surface of the microsphere to realize the PABE detection based on Cas9 / sgRNA ( Figure 4 ).
[0069] experimental method:
[0070] Selection of Cas9 / sgRNA binding sites on the HPV18 L1 gene: design two HPV18 L1 gene-specific sgRNA targets, the sequences of the two targets and their PAM sequences are: 5′-GCA TCA TAT TGC CCA GGT ACA GG- 3' (HPV18L1-1; Table 2) and 5'-AAA CCA AAT TTA TTT GGG TCA GG-3' (HPV18L1-2; Table 2). The distance between the two targets is 839bp...
Embodiment 2
[0079] Example 2 Quantitative detection of HPV18 L1 gene fragments by PABE method
[0080] experimental method:
[0081] Selection of the Cas9 / sgRNA binding site on the HPV18 L1 gene: the same as in Example 1.
[0082] Preparation of sgRNA by in vitro transcription method: same as Example 1.
[0083] Preparation of HPV L1 gene fragments by PCR: same as in Example 1.
[0084] Preparation of capture microspheres: same as Example 1.
[0085] Using dCas9 / sgRNA to combine HPV L1 gene fragments: recombinant dCas9 protein was purchased from GenScript (GeneScript, Nanjing). The dCas9 reaction (30 μL) consists of 1 × dCas9 reaction buffer (the components are the same as NEB Cas9 nuclease reaction buffer), 1 μM dCas9 (GeneScript), 300 nM sgRNA1 (HPV18L1-1; Table 2), 300 nM sgRNA (HPV18L1-2; Table 2 ), and 0.5 μL RNase inhibitor (Thermo). Incubate at room temperature for 5 minutes (this process is hereinafter referred to as pre-assembly). The above solution was mixed (32 μL) with d...
Embodiment 3
[0089] Example 3 Detection of target DNA fragments with different sgRNA target spacing by PABE method
[0090] experimental method:
[0091] Preparation of sgRNA by in vitro transcription method: sgRNA1 and sgRNA2 were prepared according to the operation method of Example 1. When preparing sgRNA1 against target DNA, use F1, R, F2, F3, and sgR1 oligonucleotides to perform three rounds of PCR amplification to prepare sgRNA1 transcription templates. When preparing sgRNA2 against target DNA, use F1, R, F2, F3, and sgR2 oligonucleotides for three rounds of PCR amplification to prepare sgRNA2 transcription templates. The PCR reaction system and reaction procedure are the same as in Example 1.
[0092] Target DNA fragment preparation: synthesize two free bases with two sgRNA target sequences and 10bp respectively. Among the primers used, the upstream primer is a fixed sequence with HPV18-E6-E7-1 at the 5′ end Target sequence (Ladder-F; Table 1); the downstream primer has the targe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com