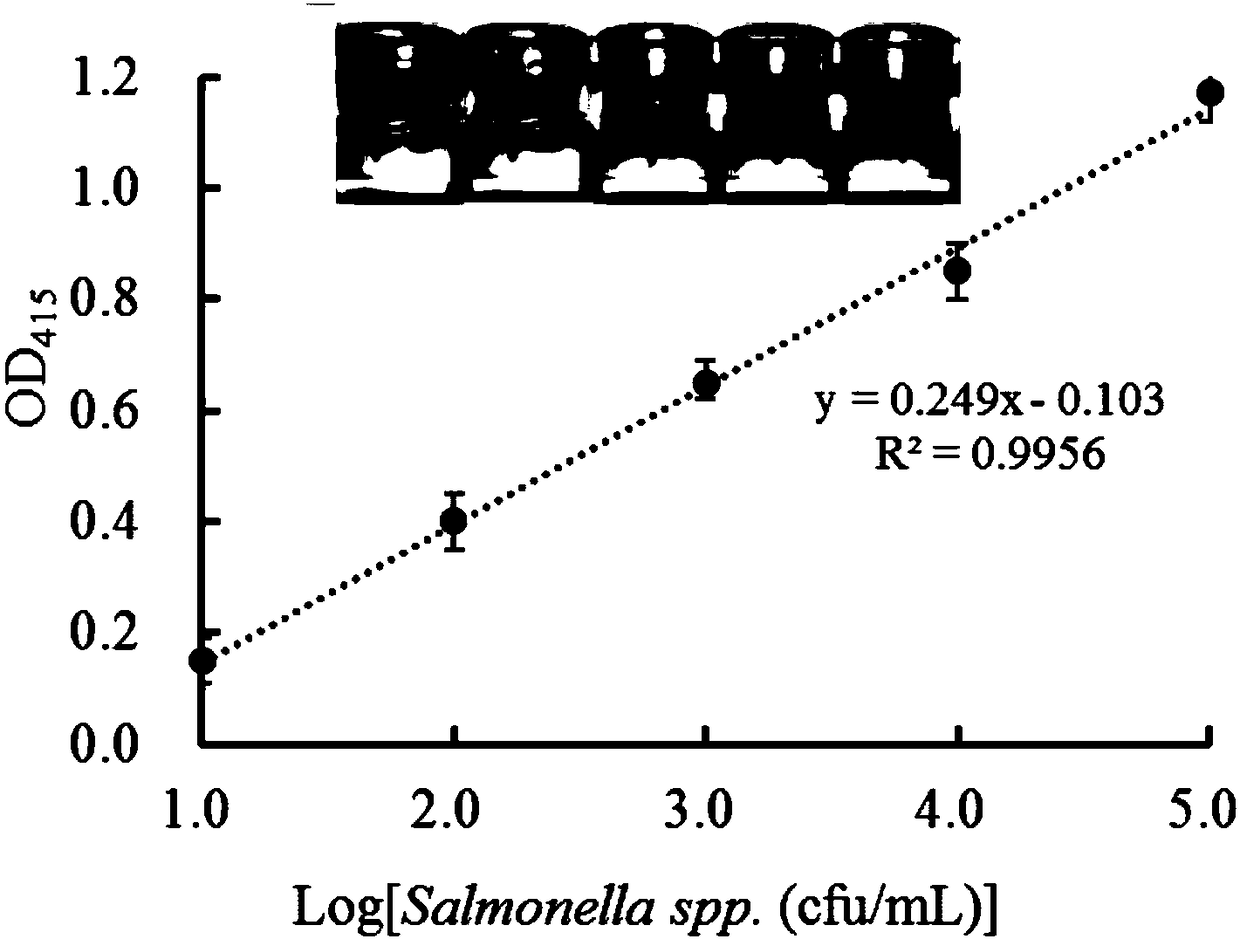
Novel colorimetric sensing method for dual pathogenic bacteria
A detection method and nucleotide sequence technology, applied in the field of biological detection, can solve the problems of difficult analysis results, cumbersome experimental operations, cumbersome operations, etc., and achieve the effect of simple and efficient detection and analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0119] Example 1. Establishment of a new ultra-fast PCR-based dual colorimetric sensing method for the detection of Salmonella and Staphylococcus aureus
[0120] (1) Experimental materials
[0121] The strain information used in this example is shown in Table 1, and the nucleotide sequences of the designed primers are shown in Table 2 and the sequence list.
[0122] Table 1
[0123]
[0124] Table 2
[0125]
[0126] In Table 2, the nucleotide sequence on the left side of the connecting arm (oxyethyleneglycol bridge) of the upstream primer Primer 1 is the nucleotide sequence shown in SEQ ID No. 1 in the sequence table, and the nucleotide sequence on the right side of the connecting arm is the sequence The nucleotide sequence shown in SEQID №: 2 in the list, the chemical structure of the connecting arm is:
[0127]
[0128] In Table 2, the nucleotide sequence of the downstream primer Primer 2 is the nucleotide sequence shown in SEQ ID No.: 3 in the sequence listing....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com