Method for genome copy number variation based on target region capturing sequencing
A technology for target region capture and copy number variation, applied in the field of genetic engineering, can solve problems such as unguaranteed accuracy and inapplicability, and achieve the effect of improving efficiency, improving accuracy and sensitivity, and reducing the demand for computing resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
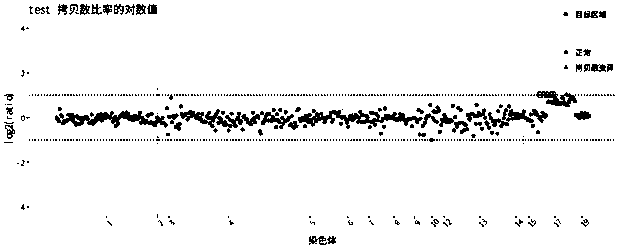
[0035] refer to Figure 1-2 , In this embodiment, 5 samples of normal people were collected to make initial data, and another sample was collected as a test sample.
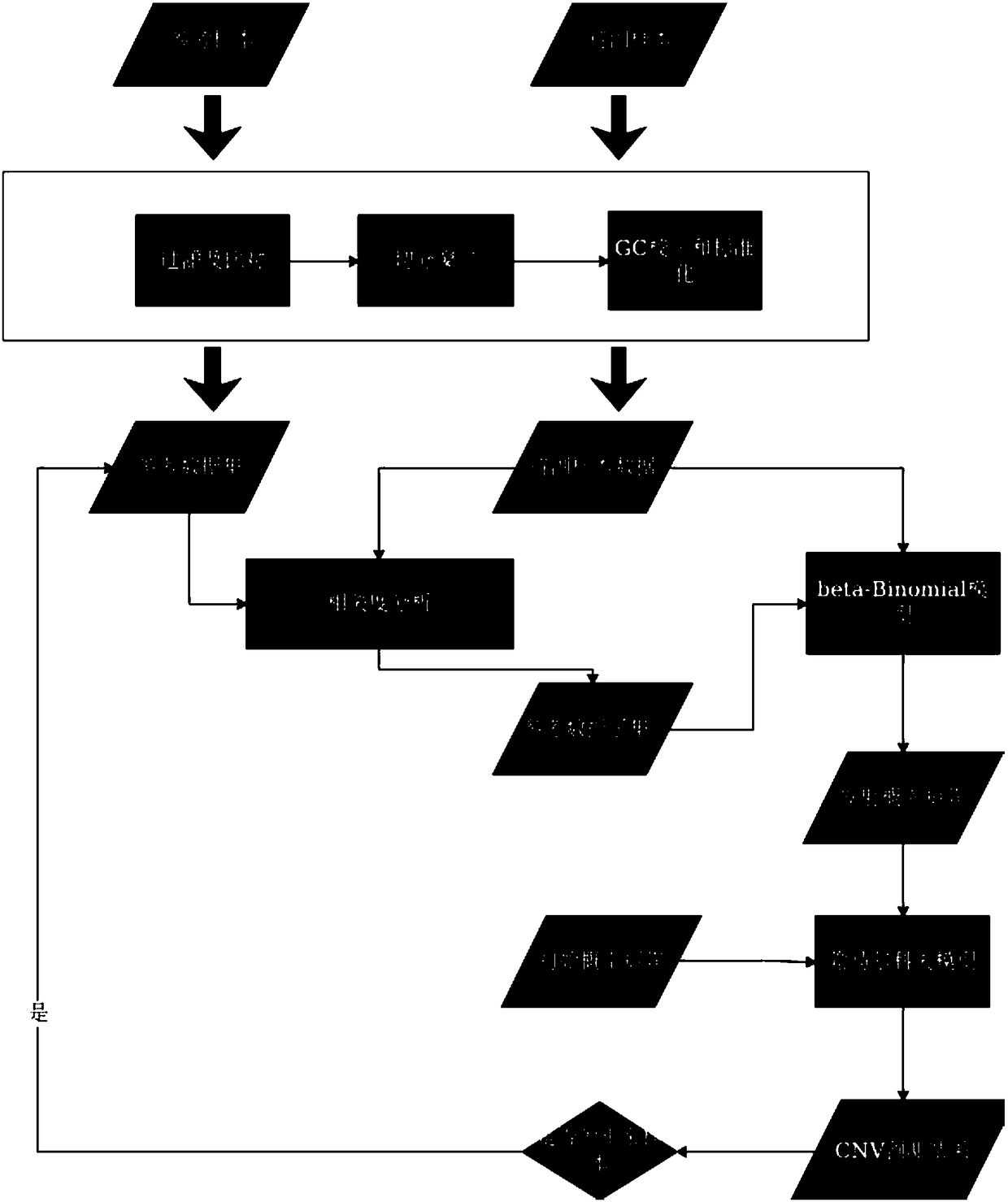
[0036] The method for capturing and sequencing genome copy number variation based on the target region in this embodiment is performed according to the following steps:
[0037] (1) Divide the window of the target area to obtain the window file. The initial setting window length is about 60, and the coincidence degree is about 20%. The following is the intercepted part of the window table:
[0038]
[0039] (2) Use the miseq platform to sequence the fastq file, remove the fragments with a quality value below 10, and remove the adapter data of the fragments, use the BWA comparison software to compare the fasq file to the human reference genome (UCSC_hg19), after the comparison is obtained bam file, where the reference genome is downloaded from the UCSC website;
[0040] (3) Count the uniquely aligned fragment...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com